Significant effects of host dietary guild and phylogeny in wild lemur gut microbiomes
- PMID: 37938265
- PMCID: PMC9723590
- DOI: 10.1038/s43705-022-00115-6
Significant effects of host dietary guild and phylogeny in wild lemur gut microbiomes
Abstract
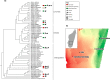
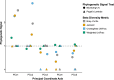
Mammals harbor diverse gut microbiomes (GMs) that perform critical functions for host health and fitness. Identifying factors associated with GM variation can help illuminate the role of microbial symbionts in mediating host ecological interactions and evolutionary processes, including diversification and adaptation. Many mammals demonstrate phylosymbiosis-a pattern in which more closely-related species harbor more similar GMs-while others show overwhelming influences of diet and habitat. Here, we generated 16S rRNA sequence data from fecal samples of 15 species of wild lemurs across southern Madagascar to (1) test a hypothesis of phylosymbiosis, and (2) test trait correlations between dietary guild, habitat, and GM diversity. Our results provide strong evidence of phylosymbiosis, though some closely-related species with substantial ecological niche overlap exhibited greater GM similarity than expected under Brownian motion. Phylogenetic regressions also showed a significant correlation between dietary guild and UniFrac diversity, but not Bray-Curtis or Jaccard. This discrepancy between beta diversity metrics suggests that older microbial clades have stronger associations with diet than younger clades, as UniFrac weights older clades more heavily. We conclude that GM diversity is predominantly shaped by host phylogeny, and that microbes associated with diet were likely acquired before evolutionary radiations within the lemur families examined.
© 2022. The Author(s).
Conflict of interest statement
Patricia C Wright is on the advisory board of Primate Conservation, Inc. (PCI), one of the funders of this project. She did not advise PCI on funding this project.
Figures

Similar articles
-
Lemur Gut Microeukaryotic Community Variation Is Not Associated with Host Phylogeny, Diet, or Habitat.Microb Ecol. 2023 Oct;86(3):2149-2160. doi: 10.1007/s00248-023-02233-7. Epub 2023 May 3. Microb Ecol. 2023. PMID: 37133496
-
Evolutionary history influences the microbiomes of a female symbiotic reproductive organ in cephalopods.Appl Environ Microbiol. 2024 Mar 20;90(3):e0099023. doi: 10.1128/aem.00990-23. Epub 2024 Feb 5. Appl Environ Microbiol. 2024. PMID: 38315021 Free PMC article.
-
Gut microbiota of ring-tailed lemurs (Lemur catta) vary across natural and captive populations and correlate with environmental microbiota.Anim Microbiome. 2022 Apr 28;4(1):29. doi: 10.1186/s42523-022-00176-x. Anim Microbiome. 2022. PMID: 35484581 Free PMC article.
-
Host specificity of the gut microbiome.Nat Rev Microbiol. 2021 Oct;19(10):639-653. doi: 10.1038/s41579-021-00562-3. Epub 2021 May 27. Nat Rev Microbiol. 2021. PMID: 34045709 Review.
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
Cited by
-
The gut microbiome of Madagascar's lemurs from forest fragments in the central highlands.Primates. 2025 May;66(3):313-325. doi: 10.1007/s10329-025-01182-8. Epub 2025 Feb 20. Primates. 2025. PMID: 39976822
-
The Role of Geography, Diet, and Host Phylogeny on the Gut Microbiome in the Hawaiian Honeycreeper Radiation.Ecol Evol. 2024 Oct 16;14(10):e70372. doi: 10.1002/ece3.70372. eCollection 2024 Oct. Ecol Evol. 2024. PMID: 39416467 Free PMC article.
-
Host genetics and larval host plant modulate microbiome structure and evolution underlying the intimate insect-microbe-plant interactions in Parnassius species on the Qinghai-Tibet Plateau.Ecol Evol. 2024 Apr 10;14(4):e11218. doi: 10.1002/ece3.11218. eCollection 2024 Apr. Ecol Evol. 2024. PMID: 38606343 Free PMC article.
-
Why Didn't the Sifaka Cross the Road? Divergence of Propithecus edwardsi Gut Microbiomes Across Geographic Barriers in Ranomafana National Park, Madagascar.Am J Primatol. 2025 Feb;87(2):e23732. doi: 10.1002/ajp.23732. Am J Primatol. 2025. PMID: 39905243 Free PMC article.

