SCFβTrCP-mediated degradation of SHARP1 in triple-negative breast cancer
- PMID: 37938564
- PMCID: PMC10632515
- DOI: 10.1038/s41419-023-06253-6
SCFβTrCP-mediated degradation of SHARP1 in triple-negative breast cancer
Abstract
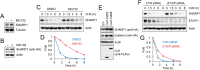
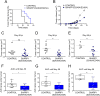
Triple-negative breast cancer (TNBC) is a subtype of breast cancer associated with metastasis, high recurrence rate, and poor survival. The basic helix-loop-helix transcription factor SHARP1 (Split and Hairy-related Protein 1) has been identified as a suppressor of the metastatic behavior of TNBC. SHARP1 blocks the invasive phenotype of TNBC by inhibiting hypoxia-inducible factors and its loss correlates with poor survival of breast cancer patients. Here, we show that SHARP1 is an unstable protein that is targeted for proteasomal degradation by the E3 ubiquitin ligase complex SCFβTrCP. SHARP1 recruits βTrCP via a phosphodegron encompassing Ser240 and Glu245 which are required for SHARP1 ubiquitylation and degradation. Furthermore, mice injected with TNBC cells expressing the non-degradable SHARP1(S240A/E245A) mutant display reduced tumor growth and increased tumor-free survival. Our study suggests that targeting the βTrCP-dependent degradation of SHARP1 represents a therapeutic strategy in TNBC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

