Drought-responsive genes in tomato: meta-analysis of gene expression using machine learning
- PMID: 37938584
- PMCID: PMC10632472
- DOI: 10.1038/s41598-023-45942-2
Drought-responsive genes in tomato: meta-analysis of gene expression using machine learning
Abstract
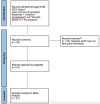
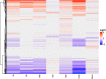
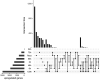
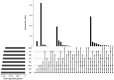
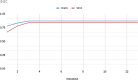
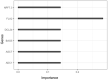
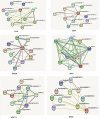
Plants have diverse molecular mechanisms to protect themselves from biotic and abiotic stressors and adapt to changing environments. To uncover the genetic potential of plants, it is crucial to understand how they adapt to adverse conditions by analyzing their genomic data. We analyzed RNA-Seq data from different tomato genotypes, tissue types, and drought durations. We used a time series scale to identify early and late drought-responsive gene modules and applied a machine learning method to identify the best responsive genes to drought. We demonstrated six candidate genes of tomato viz. Fasciclin-like arabinogalactan protein 2 (FLA2), Amino acid transporter family protein (ASCT), Arginine decarboxylase 1 (ADC1), Protein NRT1/PTR family 7.3 (NPF7.3), BAG family molecular chaperone regulator 5 (BAG5) and Dicer-like 2b (DCL2b) were responsive to drought. We constructed gene association networks to identify their potential interactors and found them drought-responsive. The identified candidate genes can help to explore the adaptation of tomato plants to drought. Furthermore, these candidate genes can have far-reaching implications for molecular breeding and genome editing in tomatoes, providing insights into the molecular mechanisms that underlie drought adaptation. This research underscores the importance of the genetic basis of plant adaptation, particularly in changing climates and growing populations.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hayat S, Hasan SA, Fariduddin Q, Ahmad A. Growth of tomato (Lycopersicon esculentum) in response to salicylic acid under water stress. J. Plant Inter. 2008;3:297–304. doi: 10.1080/17429140802320797. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

