Farm-scale differentiation of active microbial colonizers
- PMID: 37938671
- PMCID: PMC9723676
- DOI: 10.1038/s43705-022-00120-9
Farm-scale differentiation of active microbial colonizers
Abstract
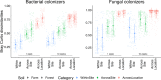
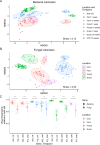
Microbial movement is important for replenishing lost soil microbial biodiversity and driving plant root colonization, particularly in managed agricultural soils, where microbial diversity and composition can be disrupted. Despite abundant survey-type microbiome data in soils, which are obscured by legacy DNA and microbial dormancy, we do not know how active microbial pools are shaped by local soil properties, agricultural management, and at differing spatial scales. To determine how active microbial colonizers are shaped by spatial scale and environmental conditions, we deployed microbial traps (i.e. sterile soil enclosed by small pore membranes) containing two distinct soil types (forest; agricultural), in three neighboring locations, assessing colonization through 16S rRNA gene and fungal ITS amplicon sequencing. Location had a greater impact on fungal colonizers (R2 = 0.31 vs. 0.26), while the soil type within the microbial traps influenced bacterial colonizers more (R2 = 0.09 vs. 0.02). Bacterial colonizers showed greater colonization consistency (within-group similarity) among replicate communities. Relative to bacterial colonizers, fungal colonizers shared a greater compositional overlap to sequences from the surrounding local bulk soil (R2 = 0.08 vs. 0.29), suggesting that these groups respond to distinct environmental constraints and that their in-field management may differ. Understanding how environmental constraints and spatial scales impact microbial recolonization dynamics and community assembly are essential for identifying how soil management can be used to shape agricultural microbiomes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Blagodatskaya E, Kuzyakov Y. Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol Biochem. 2013;67:192–211. doi: 10.1016/j.soilbio.2013.08.024. - DOI

