Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes
- PMID: 37938773
- PMCID: PMC10651487
- DOI: 10.1038/s41586-023-06708-y
Deconstruction of rheumatoid arthritis synovium defines inflammatory subtypes
Abstract
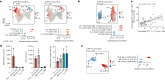
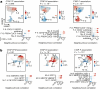
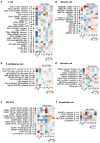
Rheumatoid arthritis is a prototypical autoimmune disease that causes joint inflammation and destruction1. There is currently no cure for rheumatoid arthritis, and the effectiveness of treatments varies across patients, suggesting an undefined pathogenic diversity1,2. Here, to deconstruct the cell states and pathways that characterize this pathogenic heterogeneity, we profiled the full spectrum of cells in inflamed synovium from patients with rheumatoid arthritis. We used multi-modal single-cell RNA-sequencing and surface protein data coupled with histology of synovial tissue from 79 donors to build single-cell atlas of rheumatoid arthritis synovial tissue that includes more than 314,000 cells. We stratified tissues into six groups, referred to as cell-type abundance phenotypes (CTAPs), each characterized by selectively enriched cell states. These CTAPs demonstrate the diversity of synovial inflammation in rheumatoid arthritis, ranging from samples enriched for T and B cells to those largely lacking lymphocytes. Disease-relevant cell states, cytokines, risk genes, histology and serology metrics are associated with particular CTAPs. CTAPs are dynamic and can predict treatment response, highlighting the clinical utility of classifying rheumatoid arthritis synovial phenotypes. This comprehensive atlas and molecular, tissue-based stratification of rheumatoid arthritis synovial tissue reveal new insights into rheumatoid arthritis pathology and heterogeneity that could inform novel targeted treatments.
© 2023. The Author(s).
Conflict of interest statement
A.H.J. reports research support from Amgen outside the submitted work. K. Wei is a consultant for Mestag Therapeutics and Gilead Sciences and reports grant support from Gilead Sciences. S.M.G. reports research support from Novartis and is a consultant for UCB, outside the submitted work. V.M.H. is a co-founder of Q32 Bio and has previously received sponsored research from Janssen and been a consultant for Celgene and BMS, outside the submitted work. A.F. reports personal fees from Abbvie, Roche and Janssen and grant support from Roche, UCB, Nascient, Mestag, GlaxoSmithKline and Janssen, outside the submitted work. D.A.R. reports personal fees from Pfizer, Janssen, Merck, Scipher Medicine, GlaxoSmithKline and Bristol-Myers Squibb and grant support from Janssen and Bristol-Myers Squibb, outside the submitted work. In addition, D.A.R. is a co-inventor on a patent submitted on T peripheral helper cells. M.B.B. is a founder for Mestag Therapeutics and a consultant for GlaxoSmithKline, 4FO Ventures and Scailyte AG. S.R. is a founder for Mestag Therapeutics, a scientific advisor for Janssen and Pfizer, and a consultant for Gilead and Rheos Medicines.
Figures












References
-
- Alivernini S, Laria A, Gremese E, Zoli A, Ferraccioli G. ACR70-disease activity score remission achievement from switches between all the available biological agents in rheumatoid arthritis: a systematic review of the literature. Arthritis Res. Ther. 2009;11:R163. doi: 10.1186/ar2848. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI148102/AI/NIAID NIH HHS/United States
- UH2 AR067685/AR/NIAMS NIH HHS/United States
- UH2 AR067681/AR/NIAMS NIH HHS/United States
- UH2 AR067694/AR/NIAMS NIH HHS/United States
- K08 AR077037/AR/NIAMS NIH HHS/United States
- R01 AI148435/AI/NIAID NIH HHS/United States
- UH2 AR067679/AR/NIAMS NIH HHS/United States
- UM2 AR067678/AR/NIAMS NIH HHS/United States
- P30 AR069655/AR/NIAMS NIH HHS/United States
- R21 AR071670/AR/NIAMS NIH HHS/United States
- K08 AR081412/AR/NIAMS NIH HHS/United States
- T32 AR007258/AR/NIAMS NIH HHS/United States
- MR/M007669/1/MRC_/Medical Research Council/United Kingdom
- UL1 TR001866/TR/NCATS NIH HHS/United States
- P30 AR070253/AR/NIAMS NIH HHS/United States
- R01 AR073290/AR/NIAMS NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- T32 AR007530/AR/NIAMS NIH HHS/United States
- MR/P021220/1/MRC_/Medical Research Council/United Kingdom
- R01 AR073833/AR/NIAMS NIH HHS/United States
- UH2 AR067688/AR/NIAMS NIH HHS/United States
- K08 AR072791/AR/NIAMS NIH HHS/United States
- T32 GM144273/GM/NIGMS NIH HHS/United States
- UH2 AR067689/AR/NIAMS NIH HHS/United States
- R01 AR063759/AR/NIAMS NIH HHS/United States
- UH2 AR067690/AR/NIAMS NIH HHS/United States
- U01 HG009379/HG/NHGRI NIH HHS/United States
- UH2 AR067677/AR/NIAMS NIH HHS/United States
- UH2 AR067676/AR/NIAMS NIH HHS/United States
- UH2 AR067691/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

