Pioneer factors: roles and their regulation in development
- PMID: 37940484
- PMCID: PMC10873006
- DOI: 10.1016/j.tig.2023.10.007
Pioneer factors: roles and their regulation in development
Abstract
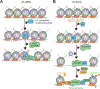
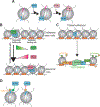
Pioneer factors are a subclass of transcription factors that can bind and initiate opening of silent chromatin regions. Pioneer factors subsequently regulate lineage-specific genes and enhancers and, thus, activate the zygotic genome after fertilization, guide cell fate transitions during development, and promote various forms of human cancers. As such, pioneer factors are useful in directed cell reprogramming. In this review, we define the structural and functional characteristics of pioneer factors, how they bind and initiate opening of closed chromatin regions, and the consequences for chromatin dynamics and gene expression during cell differentiation. We also discuss emerging mechanisms that modulate pioneer factors during development.
Keywords: development; gene expression; heterochromatin; nucleosome; pioneer factor; zygotic gene activation.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures



References
-
- Boyer LA et al. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441 (7091), 349–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

