Exome sequencing (ES) of a pediatric cohort with chronic endocrine diseases: a single-center study (within the framework of the TRANSLATE-NAMSE project)
- PMID: 37940764
- PMCID: PMC11246252
- DOI: 10.1007/s12020-023-03581-7
Exome sequencing (ES) of a pediatric cohort with chronic endocrine diseases: a single-center study (within the framework of the TRANSLATE-NAMSE project)
Abstract
Background: Endocrine disorders are heterogeneous and include a significant number of rare monogenic diseases.
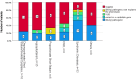
Methods: We performed exome sequencing (ES) in 106 children recruited from a single center within the TRANSLATE‑NAMSE project. They were categorized into subgroups: proportionate short stature (PSS), disproportionate short stature (DSS), hypopituitarism (H), differences in sexual development (DSD), syndromic diseases (SD) and others.
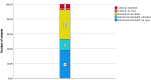
Results: The overall diagnostic yield was 34.9% (n = 37/106), including 5 patients with variants in candidate genes, which have contributed to collaborations to identify gene-disease associations. The diagnostic yield varied significantly between subgroups: PSS: 16.6% (1/6); DSS: 18.8% (3/16); H: 17.1% (6/35); DSD: 37.5% (3/8); SD: 66.6% (22/33); others: 25% (2/8). Confirmed diagnoses included 75% ultrarare diseases. Three patients harbored more than one disease-causing variant, resulting in dual diagnoses.
Conclusions: ES is an effective tool for genetic diagnosis in pediatric patients with complex endocrine diseases. An accurate phenotypic description, including comprehensive endocrine diagnostics, as well as the evaluation of variants in multidisciplinary case conferences involving geneticists, are necessary for personalized diagnostic care. Here, we illustrate the broad spectrum of genetic endocrinopathies that have led to the initiation of specific treatment, surveillance, and family counseling.
Keywords: Exome sequencing; TRANSLATE-NAMSE; chronic pediatric endocrine diseases; multidisciplinary case conferences; rare diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors have no competing interests.
Figures
References
-
- Shlomo M., Auchus R.J., Goldfine A.B., Koenig R.J., Rosen C.J. Williams textbook of endocrinology. Philadelphia: Elsevier; 2020.
-
- Bundesausschuss IbG: TRANSLATE-NAMSE – Verbesserung der Versorgung von Menschen mit seltenen Erkrankungen durch Umsetzung von im nationalen Aktionsplan (NAMSE) konsentierten Maßnahmen. https://innovationsfonds.g-ba.de/projekte/neue-versorgungsformen/transla... (2017). Accessed 20.01.2023.
-
- Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet Med. 2020;22(2):245–257. doi: 10.1038/s41436-019-0686-8. - DOI - PMC - PubMed
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

