The Reactome Pathway Knowledgebase 2024
- PMID: 37941124
- PMCID: PMC10767911
- DOI: 10.1093/nar/gkad1025
The Reactome Pathway Knowledgebase 2024
Abstract
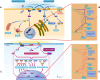
The Reactome Knowledgebase (https://reactome.org), an Elixir and GCBR core biological data resource, provides manually curated molecular details of a broad range of normal and disease-related biological processes. Processes are annotated as an ordered network of molecular transformations in a single consistent data model. Reactome thus functions both as a digital archive of manually curated human biological processes and as a tool for discovering functional relationships in data such as gene expression profiles or somatic mutation catalogs from tumor cells. Here we review progress towards annotation of the entire human proteome, targeted annotation of disease-causing genetic variants of proteins and of small-molecule drugs in a pathway context, and towards supporting explicit annotation of cell- and tissue-specific pathways. Finally, we briefly discuss issues involved in making Reactome more fully interoperable with other related resources such as the Gene Ontology and maintaining the resulting community resource network.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

