WikiPathways 2024: next generation pathway database
- PMID: 37941138
- PMCID: PMC10767877
- DOI: 10.1093/nar/gkad960
WikiPathways 2024: next generation pathway database
Abstract
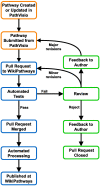
WikiPathways (wikipathways.org) is an open-source biological pathway database. Collaboration and open science are pivotal to the success of WikiPathways. Here we highlight the continuing efforts supporting WikiPathways, content growth and collaboration among pathway researchers. As an evolving database, there is a growing need for WikiPathways to address and overcome technical challenges. In this direction, WikiPathways has undergone major restructuring, enabling a renewed approach for sharing and curating pathway knowledge, thus providing stability for the future of community pathway curation. The website has been redesigned to improve and enhance user experience. This next generation of WikiPathways continues to support existing features while improving maintainability of the database and facilitating community input by providing new functionality and leveraging automation.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

