A conserved requirement for RME-8/DNAJC13 in neuronal autophagic lysosome reformation
- PMID: 37942902
- PMCID: PMC11062384
- DOI: 10.1080/15548627.2023.2269028
A conserved requirement for RME-8/DNAJC13 in neuronal autophagic lysosome reformation
Abstract
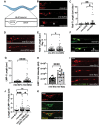
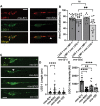
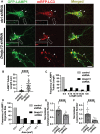
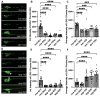
Autophagosomes fuse with lysosomes, forming autolysosomes that degrade engulfed cargo. To maintain lysosomal capacity, autophagic lysosome reformation (ALR) must regenerate lysosomes from autolysosomes using a membrane tubule-based process. Maintaining lysosomal capacity is required to maintain cellular health, especially in neurons where lysosomal dysfunction has been repeatedly implicated in neurodegenerative disease. The DNA-J domain HSC70 co-chaperone RME-8/DNAJC13 has been linked to endosomal coat protein regulation and to neurological disease. We report new analysis of the requirements for the RME-8/DNAJC13 protein in neurons, focusing on intact C. elegans mechanosensory neurons, and primary mouse cortical neurons in culture. Loss of RME-8/DNAJC13 in both systems results in accumulation of grossly elongated autolysosomal tubules. Further C. elegans analysis revealed a similar autolysosome tubule accumulation defect in mutants known to be required for ALR in mammals, including mutants lacking bec-1/BECN1/Beclin1 and vps-15/PIK3R4/p150 that regulate the class III phosphatidylinositol 3-kinase (PtdIns3K) VPS-34, and dyn-1/dynamin that severs ALR tubules. Clathrin is also an important ALR regulator implicated in autolysosome tubule formation and release. In C. elegans we found that loss of RME-8 causes severe depletion of clathrin from neuronal autolysosomes, a phenotype shared with bec-1 and vps-15 mutants. We conclude that RME-8/DNAJC13 plays a previously unrecognized role in ALR, likely affecting autolysosome tubule severing. Additionally, in both systems, loss of RME-8/DNAJC13 reduced macroautophagic/autophagic flux, suggesting feedback regulation from ALR to autophagy. Our results connecting RME-8/DNAJC13 to ALR and autophagy provide a potential mechanism by which RME-8/DNAJC13 could influence neuronal health and the progression of neurodegenerative disease.Abbreviation: ALR, autophagic lysosome reformation; ATG-13/EPG-1, AuTophaGy (yeast Atg homolog)-13; ATG-18, AuTophaGy (yeast Atg homolog)-18; AV, autophagic vacuole; CLIC-1, Clathrin Light Chain-1; EPG-3, Ectopic P Granules-3; EPG-6, Ectopic P Granules-6; LGG-1, LC3, GABARAP and GATE-16 family-1; MAP1LC3/LC3, microtubule-associated protein 1 light chain 3; PD, Parkinson disease; PtdIns3P, phosphatidylinositol-3-phosphate; PtdIns(4,5)P2, phosphatidylinositol-4,5-bisphosphate; RME-8, Receptor Mediated Endocytosis-8; SNX-1, Sorting NeXin-1; VPS-34, related to yeast Vacuolar Protein Sorting factor-34.
Keywords: Autophagy; clathrin; endocytosis; lysosomes; neurodegeneration; trafficking.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



Update of
-
A Conserved Requirement for RME-8/DNAJC13 in Neuronal Autolysosome Reformation.bioRxiv [Preprint]. 2023 Feb 28:2023.02.27.530319. doi: 10.1101/2023.02.27.530319. bioRxiv. 2023. Update in: Autophagy. 2024 Apr;20(4):792-808. doi: 10.1080/15548627.2023.2269028. PMID: 36909501 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
