COVID-19: A complex disease with a unique metabolic signature
- PMID: 37943960
- PMCID: PMC10662774
- DOI: 10.1371/journal.ppat.1011787
COVID-19: A complex disease with a unique metabolic signature
Abstract
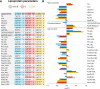
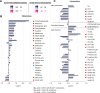
Plasma of COVID-19 patients contains a strong metabolomic/lipoproteomic signature, revealed by the NMR analysis of a cohort of >500 patients sampled during various waves of COVID-19 infection, corresponding to the spread of different variants, and having different vaccination status. This composite signature highlights common traits of the SARS-CoV-2 infection. The most dysregulated molecules display concentration trends that scale with disease severity and might serve as prognostic markers for fatal events. Metabolomics evidence is then used as input data for a sex-specific multi-organ metabolic model. This reconstruction provides a comprehensive view of the impact of COVID-19 on the entire human metabolism. The human (male and female) metabolic network is strongly impacted by the disease to an extent dictated by its severity. A marked metabolic reprogramming at the level of many organs indicates an increase in the generic energetic demand of the organism following infection. Sex-specific modulation of immune response is also suggested.
Copyright: © 2023 Ghini et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

