Clinical features, functional consequences, and rescue pharmacology of missense GRID1 and GRID2 human variants
- PMID: 37944084
- PMCID: PMC10840383
- DOI: 10.1093/hmg/ddad188
Clinical features, functional consequences, and rescue pharmacology of missense GRID1 and GRID2 human variants
Abstract
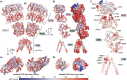
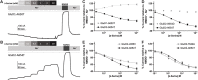
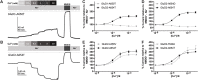
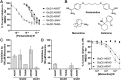
GRID1 and GRID2 encode the enigmatic GluD1 and GluD2 proteins, which form tetrameric receptors that play important roles in synapse organization and development of the central nervous system. Variation in these genes has been implicated in neurodevelopmental phenotypes. We evaluated GRID1 and GRID2 human variants from the literature, ClinVar, and clinical laboratories and found that many of these variants reside in intolerant domains, including the amino terminal domain of both GRID1 and GRID2. Other conserved regions, such as the M3 transmembrane domain, show different intolerance between GRID1 and GRID2. We introduced these variants into GluD1 and GluD2 cDNA and performed electrophysiological and biochemical assays to investigate the mechanisms of dysfunction of GRID1/2 variants. One variant in the GRID1 distal amino terminal domain resides at a position predicted to interact with Cbln2/Cbln4, and the variant disrupts complex formation between GluD1 and Cbln2, which could perturb its role in synapse organization. We also discovered that, like the lurcher mutation (GluD2-A654T), other rare variants in the GRID2 M3 domain create constitutively active receptors that share similar pathogenic phenotypes. We also found that the SCHEMA schizophrenia M3 variant GluD1-A650T produced constitutively active receptors. We tested a variety of compounds for their ability to inhibit constitutive currents of GluD receptor variants and found that pentamidine potently inhibited GluD2-T649A constitutive channels (IC50 50 nM). These results identify regions of intolerance to variation in the GRID genes, illustrate the functional consequences of GRID1 and GRID2 variants, and suggest how these receptors function normally and in disease.
Keywords: GRID1; GRID2; Cbln2; Cbln4; GluD1; GluD2; cerebellar ataxia; cerebellar atrophy; delta receptors; lurcher; schizophrenia.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
S.F.T. is a member of the SAB for Eumentis Therapeutics, Sage Therapeutics, and Combined Brain, is a member of the Medical Advisory Board for the GRIN2B Foundation and the CureGRIN Foundation, is an advisor to GRIN Therapeutics and Neurocrine, is co-founder of NeurOp Inc. and Agrithera Inc., is a member of the Board of Directors of NeurOp Inc., and is co-inventor on IP owned by Emory University. H.Y. is PI of a grant to Emory University from Sage Therapeutics. S.J.M. is PI of a grant from GRIN Therapeutics to Emory University.
Figures



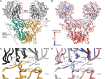



References
-
- Nakamoto C, Konno K, Miyazaki T. et al. Expression mapping, quantification, and complex formation of GluD1 and GluD2 glutamate receptors in adult mouse brain. J Comp Neurol 2020;528:1003–27. - PubMed
-
- Lomeli H, Sprengel R, Laurie DJ. et al. The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett 1993;315:318–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM137951/GM/NIGMS NIH HHS/United States
- RF1 AG081401/AG/NIA NIH HHS/United States
- U01 CA214354/CA/NCI NIH HHS/United States
- R01GM122489/NH/NIH HHS/United States
- R01 AI124680/AI/NIAID NIH HHS/United States
- GM137951/GM/NIGMS NIH HHS/United States
- R35 NS111619/NS/NINDS NIH HHS/United States
- TR003380/NS/NINDS NIH HHS/United States
- U01 CA250040/CA/NCI NIH HHS/United States
- R01 AG075444/AG/NIA NIH HHS/United States
- R03 TR003380/TR/NCATS NIH HHS/United States
- R01 AG079956/AG/NIA NIH HHS/United States
- R01 HD082373/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

