Standardized In Vitro Models of Human Adipose Tissue Reveal Metabolic Flexibility in Brown Adipocyte Thermogenesis
- PMID: 37944134
- PMCID: PMC11032247
- DOI: 10.1210/endocr/bqad161
Standardized In Vitro Models of Human Adipose Tissue Reveal Metabolic Flexibility in Brown Adipocyte Thermogenesis
Abstract
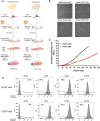
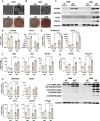
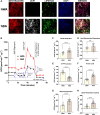
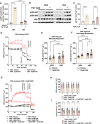
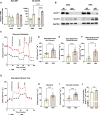
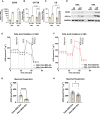
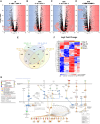
Functional human brown and white adipose tissue (BAT and WAT) are vital for thermoregulation and nutritional homeostasis, while obesity and other stressors lead, respectively, to cold intolerance and metabolic disease. Understanding BAT and WAT physiology and dysfunction necessitates clinical trials complemented by mechanistic experiments at the cellular level. These require standardized in vitro models, currently lacking, that establish references for gene expression and function. We generated and characterized a pair of immortalized, clonal human brown (hBA) and white (hWA) preadipocytes derived from the perirenal and subcutaneous depots, respectively, of a 40-year-old male individual. Cells were immortalized with hTERT and confirmed to be of a mesenchymal, nonhematopoietic lineage based on fluorescence-activated cell sorting and DNA barcoding. Functional assessments showed that the hWA and hBA phenocopied primary adipocytes in terms of adrenergic signaling, lipolysis, and thermogenesis. Compared to hWA, hBA were metabolically distinct, with higher rates of glucose uptake and lactate metabolism, and greater basal, maximal, and nonmitochondrial respiration, providing a mechanistic explanation for the association between obesity and BAT dysfunction. The hBA also responded to the stress of maximal respiration by using both endogenous and exogenous fatty acids. In contrast to certain mouse models, hBA adrenergic thermogenesis was mediated by several mechanisms, not principally via uncoupling protein 1 (UCP1). Transcriptomics via RNA-seq were consistent with the functional studies and established a molecular signature for each cell type before and after differentiation. These standardized cells are anticipated to become a common resource for future physiological, pharmacological, and genetic studies of human adipocytes.
Keywords: bioenergetics; bioinformatics; brown adipose tissue; in vitro model; white adipose tissue.
Published by Oxford University Press on behalf of the Endocrine Society 2023.
Figures
References
-
- Richard AJ, White U, Elks CM, et al. Adipose tissue: physiology to metabolic dysfunction. In: Feingold KR Anawalt B and Boyce A (eds.), Endotext. MDText.com, Inc.; 2000. - PubMed
-
- Cannon B, de Jong JMA, Fischer AW, Nedergaard J, Petrovic N. Human brown adipose tissue: classical brown rather than brite/beige? Exp Physiol. 2020;105(8):1191‐1200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

