RAGE engagement by SARS-CoV-2 enables monocyte infection and underlies COVID-19 severity
- PMID: 37944530
- PMCID: PMC10694673
- DOI: 10.1016/j.xcrm.2023.101266
RAGE engagement by SARS-CoV-2 enables monocyte infection and underlies COVID-19 severity
Abstract
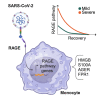
The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has fueled the COVID-19 pandemic with its enduring medical and socioeconomic challenges because of subsequent waves and long-term consequences of great concern. Here, we chart the molecular basis of COVID-19 pathogenesis by analyzing patients' immune responses at single-cell resolution across disease course and severity. This approach confirms cell subpopulation-specific dysregulation in COVID-19 across disease course and severity and identifies a severity-associated activation of the receptor for advanced glycation endproducts (RAGE) pathway in monocytes. In vitro THP1-based experiments indicate that monocytes bind the SARS-CoV-2 S1-receptor binding domain (RBD) via RAGE, pointing to RAGE-Spike interaction enabling monocyte infection. Thus, our results demonstrate that RAGE is a functional receptor of SARS-CoV-2 contributing to COVID-19 severity.
Keywords: COVID-19; RAGE; SARS-CoV-2; disease severity; drug repurposing; longitudinal profiling; monocytes; single-cell multi-omics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

