trRosettaRNA: automated prediction of RNA 3D structure with transformer network
- PMID: 37945552
- PMCID: PMC10636060
- DOI: 10.1038/s41467-023-42528-4
trRosettaRNA: automated prediction of RNA 3D structure with transformer network
Abstract
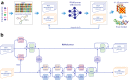
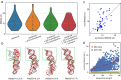
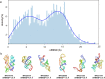
RNA 3D structure prediction is a long-standing challenge. Inspired by the recent breakthrough in protein structure prediction, we developed trRosettaRNA, an automated deep learning-based approach to RNA 3D structure prediction. The trRosettaRNA pipeline comprises two major steps: 1D and 2D geometries prediction by a transformer network; and 3D structure folding by energy minimization. Benchmark tests suggest that trRosettaRNA outperforms traditional automated methods. In the blind tests of the 15th Critical Assessment of Structure Prediction (CASP15) and the RNA-Puzzles experiments, the automated trRosettaRNA predictions for the natural RNAs are competitive with the top human predictions. trRosettaRNA also outperforms other deep learning-based methods in CASP15 when measured by the Z-score of the Root-Mean-Square Deviation. Nevertheless, it remains challenging to predict accurate structures for synthetic RNAs with an automated approach. We hope this work could be a good start toward solving the hard problem of RNA structure prediction with deep learning.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

