Enduring questions in regenerative biology and the search for answers
- PMID: 37945686
- PMCID: PMC10636051
- DOI: 10.1038/s42003-023-05505-7
Enduring questions in regenerative biology and the search for answers
Abstract
The potential for basic research to uncover the inner workings of regenerative processes and produce meaningful medical therapies has inspired scientists, clinicians, and patients for hundreds of years. Decades of studies using a handful of highly regenerative model organisms have significantly advanced our knowledge of key cell types and molecular pathways involved in regeneration. However, many questions remain about how regenerative processes unfold in regeneration-competent species, how they are curtailed in non-regenerative organisms, and how they might be induced (or restored) in humans. Recent technological advances in genomics, molecular biology, computer science, bioengineering, and stem cell research hold promise to collectively provide new experimental evidence for how different organisms accomplish the process of regeneration. In theory, this new evidence should inform the design of new clinical approaches for regenerative medicine. A deeper understanding of how tissues and organs regenerate will also undoubtedly impact many adjacent scientific fields. To best apply and adapt these new technologies in ways that break long-standing barriers and answer critical questions about regeneration, we must combine the deep knowledge of developmental and evolutionary biologists with the hard-earned expertise of scientists in mechanistic and technical fields. To this end, this perspective is based on conversations from a workshop we organized at the Banbury Center, during which a diverse cross-section of the regeneration research community and experts in various technologies discussed enduring questions in regenerative biology. Here, we share the questions this group identified as significant and unanswered, i.e., known unknowns. We also describe the obstacles limiting our progress in answering these questions and how expanding the number and diversity of organisms used in regeneration research is essential for deepening our understanding of regenerative capacity. Finally, we propose that investigating these problems collaboratively across a diverse network of researchers has the potential to advance our field and produce unexpected insights into important questions in related areas of biology and medicine.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
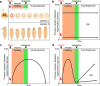
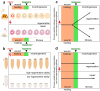

References
-
- Kuhn, T. S. The structure of scientific revolutions. 2nd edn. (Chicago University Press, 1970).
-
- Needham, A. E. Regeneration and wound-healing. (J. Wiley, 1952).
-
- Goldstein, B. & Srivastava, M. Emerging model systems in developmental biology. First edition. (Elsevier/Academic Press, 2022).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

