Regulatory T cell-derived IL-1Ra suppresses the innate response to respiratory viral infection
- PMID: 37945820
- PMCID: PMC11887468
- DOI: 10.1038/s41590-023-01655-2
Regulatory T cell-derived IL-1Ra suppresses the innate response to respiratory viral infection
Abstract
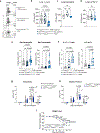
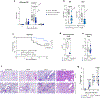
Regulatory T (Treg) cell modulation of adaptive immunity and tissue homeostasis is well described; however, less is known about Treg cell-mediated regulation of the innate immune response. Here we show that deletion of ST2, the receptor for interleukin (IL)-33, on Treg cells increased granulocyte influx into the lung and increased cytokine production by innate lymphoid and γδ T cells without alteration of adaptive immunity to influenza. IL-33 induced high levels of the interleukin-1 receptor antagonist (IL-1Ra) in ST2+ Treg cells and deletion of IL-1Ra in Treg cells increased granulocyte influx into the lung. Treg cell-specific deletion of ST2 or IL-1Ra improved survival to influenza, which was dependent on IL-1. Adventitial fibroblasts in the lung expressed high levels of the IL-1 receptor and their chemokine production was suppressed by Treg cell-produced IL-1Ra. Thus, we define a new pathway where IL-33-induced IL-1Ra production by tissue Treg cells suppresses IL-1-mediated innate immune responses to respiratory viral infection.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures















References
Methods References:
-
- Itohara S. et al. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell 72, 337–348 (1993). - PubMed
-
- Lamacchia C, Palmer G, Seemayer CA, Talabot-Ayer D. & Gabay C. Enhanced Th1 and Th17 responses and arthritis severity in mice with a deficiency of myeloid cell-specific interleukin-1 receptor antagonist. Arthritis Rheum 62, 452–462 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

