Circular extrachromosomal DNA promotes tumor heterogeneity in high-risk medulloblastoma
- PMID: 37945900
- PMCID: PMC10703696
- DOI: 10.1038/s41588-023-01551-3
Circular extrachromosomal DNA promotes tumor heterogeneity in high-risk medulloblastoma
Abstract
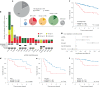
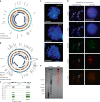
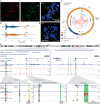
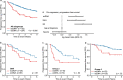
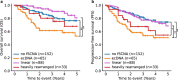
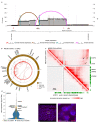
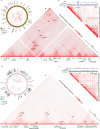
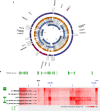
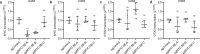
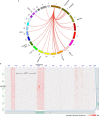
Circular extrachromosomal DNA (ecDNA) in patient tumors is an important driver of oncogenic gene expression, evolution of drug resistance and poor patient outcomes. Applying computational methods for the detection and reconstruction of ecDNA across a retrospective cohort of 481 medulloblastoma tumors from 465 patients, we identify circular ecDNA in 82 patients (18%). Patients with ecDNA-positive medulloblastoma were more than twice as likely to relapse and three times as likely to die within 5 years of diagnosis. A subset of tumors harbored multiple ecDNA lineages, each containing distinct amplified oncogenes. Multimodal sequencing, imaging and CRISPR inhibition experiments in medulloblastoma models reveal intratumoral heterogeneity of ecDNA copy number per cell and frequent putative 'enhancer rewiring' events on ecDNA. This study reveals the frequency and diversity of ecDNA in medulloblastoma, stratified into molecular subgroups, and suggests copy number heterogeneity and enhancer rewiring as oncogenic features of ecDNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: H.Y.C. is a co-founder of Accent Therapeutics, Boundless Bio, Cartography Biosciences, Orbital Therapeutics and is an advisor of 10x Genomics, Arsenal Biosciences, Chroma Medicine and Spring Discovery. P.S.M. is a co-founder, chairs the scientific advisory board (SAB) and has equity interest in Boundless Bio. P.S.M. is also an advisor with equity for Asteroid Therapeutics and is an advisor to Sage Therapeutics. V.B. is a co-founder, consultant, SAB member and has equity interest in Boundless Bio and Abterra. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. J.L. is a part-time consultant for Boundless Bio. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. J.T.L. is an employee of Boundless Bio. His employment began after his contributions to the manuscript. The remaining authors declare no competing interests.
Figures



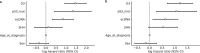

Comment in
-
Defining the Role of Extrachromosomal DNA Amplifications in Medulloblastoma.Cancer Res. 2024 Feb 15;84(4):515-516. doi: 10.1158/0008-5472.CAN-23-4025. Cancer Res. 2024. PMID: 38175761
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM074024/GM/NIGMS NIH HHS/United States
- R01 GM132292/GM/NIGMS NIH HHS/United States
- F99 CA274692/CA/NCI NIH HHS/United States
- R01 CA238379/CA/NCI NIH HHS/United States
- R01 CA159859/CA/NCI NIH HHS/United States
- CGCATF-2021/100012/CRUK_/Cancer Research UK/United Kingdom
- T15 LM011271/LM/NLM NIH HHS/United States
- U01 CA184898/CA/NCI NIH HHS/United States
- U24 CA220341/CA/NCI NIH HHS/United States
- OT2 CA278635/CA/NCI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R01 GM114362/GM/NIGMS NIH HHS/United States
- R21 NS120075/NS/NINDS NIH HHS/United States
- OT2 CA278688/CA/NCI NIH HHS/United States
- U24 CA210004/CA/NCI NIH HHS/United States
- R21 NS116455/NS/NINDS NIH HHS/United States
- U24 CA258406/CA/NCI NIH HHS/United States
- R35 NS122339/NS/NINDS NIH HHS/United States
- R01 CA238249/CA/NCI NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- U01 CA253547/CA/NCI NIH HHS/United States
- R01 CA262746/CA/NCI NIH HHS/United States
- U24 CA248457/CA/NCI NIH HHS/United States
- U24 CA264379/CA/NCI NIH HHS/United States
- F31 CA271777/CA/NCI NIH HHS/United States
- R21 NS130137/NS/NINDS NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- R01 NS132780/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

