Spatial and single-cell profiling of the metabolome, transcriptome and epigenome of the aging mouse liver
- PMID: 37946043
- PMCID: PMC10645594
- DOI: 10.1038/s43587-023-00513-y
Spatial and single-cell profiling of the metabolome, transcriptome and epigenome of the aging mouse liver
Abstract
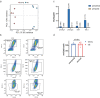
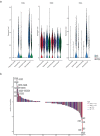
Tissues within an organism and even cell types within a tissue can age with different velocities. However, it is unclear whether cells of one type experience different aging trajectories within a tissue depending on their spatial location. Here, we used spatial transcriptomics in combination with single-cell ATAC-seq and RNA-seq, lipidomics and functional assays to address how cells in the male murine liver are affected by age-related changes in the microenvironment. Integration of the datasets revealed zonation-specific and age-related changes in metabolic states, the epigenome and transcriptome. The epigenome changed in a zonation-dependent manner and functionally, periportal hepatocytes were characterized by decreased mitochondrial fitness, whereas pericentral hepatocytes accumulated large lipid droplets. Together, we provide evidence that changing microenvironments within a tissue exert strong influences on their resident cells that can shape epigenetic, metabolic and phenotypic outputs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures







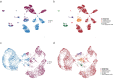




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

