Vitamin B5 supports MYC oncogenic metabolism and tumor progression in breast cancer
- PMID: 37946084
- PMCID: PMC10663155
- DOI: 10.1038/s42255-023-00915-7
Vitamin B5 supports MYC oncogenic metabolism and tumor progression in breast cancer
Abstract
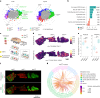
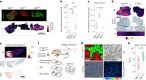
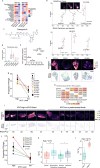
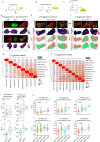
Tumors are intrinsically heterogeneous and it is well established that this directs their evolution, hinders their classification and frustrates therapy1-3. Consequently, spatially resolved omics-level analyses are gaining traction4-9. Despite considerable therapeutic interest, tumor metabolism has been lagging behind this development and there is a paucity of data regarding its spatial organization. To address this shortcoming, we set out to study the local metabolic effects of the oncogene c-MYC, a pleiotropic transcription factor that accumulates with tumor progression and influences metabolism10,11. Through correlative mass spectrometry imaging, we show that pantothenic acid (vitamin B5) associates with MYC-high areas within both human and murine mammary tumors, where its conversion to coenzyme A fuels Krebs cycle activity. Mechanistically, we show that this is accomplished by MYC-mediated upregulation of its multivitamin transporter SLC5A6. Notably, we show that SLC5A6 over-expression alone can induce increased cell growth and a shift toward biosynthesis, whereas conversely, dietary restriction of pantothenic acid leads to a reversal of many MYC-mediated metabolic changes and results in hampered tumor growth. Our work thus establishes the availability of vitamins and cofactors as a potential bottleneck in tumor progression, which can be exploited therapeutically. Overall, we show that a spatial understanding of local metabolism facilitates the identification of clinically relevant, tractable metabolic targets.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

