The oral bacterial microbiota facilitates the stratification for ulcerative colitis patients with oral ulcers
- PMID: 37946238
- PMCID: PMC10633958
- DOI: 10.1186/s12941-023-00646-3
The oral bacterial microbiota facilitates the stratification for ulcerative colitis patients with oral ulcers
Abstract
Background: Clinically, a large part of inflammatory bowel disease (IBD) patients is complicated by oral lesions. Although previous studies proved oral microbial dysbiosis in IBD patients, the bacterial community in the gastrointestinal (GI) tract of those IBD patients combined with oral ulcers has not been profiled yet.
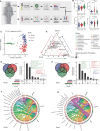
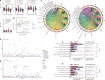
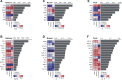
Methods: In this study, we enrolled four groups of subjects, including healthy controls (CON), oral ulcer patients (OU), and ulcerative colitis patients with (UC_OU) and without (UC) oral ulcers. Bio-samples from three GI niches containing salivary, buccal, and fecal samples, were collected for 16S rRNA V3-V4 region sequencing. Bacterial abundance and related bio-functions were compared, and data showed that the fecal microbiota was more potent than salivary and buccal microbes in shaping the host immune system. ~ 22 UC and 10 UC_OU 5-aminosalicylate (5-ASA) routine treated patients were followed-up for six months; according to their treatment response (a decrease in the endoscopic Mayo score), they were further sub-grouped as responding and non-responding patients.
Results: We found those UC patients complicated with oral ulcers presented weaker treatment response, and three oral bacterial genera, i.e., Fusobacterium, Oribacterium, and Campylobacter, might be connected with treatment responding. Additionally, the salivary microbiome could be an indicator of treatment responding in 5-ASA routine treatment rather than buccal or fecal ones.
Conclusions: The fecal microbiota had a strong effect on the host's immune indices, while the oral bacterial microbiota could help stratification for ulcerative colitis patients with oral ulcers. Additionally, the oral microbiota had the potential role in reflecting the treatment response of UC patients. Three oral bacteria genera (Fusobacterium, Oribacterium, and Campylobacter) might be involved in UC patients with oral ulcers lacking treatment responses, and monitoring oral microbiota may be meaningful in assessing the therapeutic response in UC patients.
Keywords: Bacterial microbiota; Oral ulcer; Response; Stratification; Ulcerative colitis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
The oral-gut axis: Salivary and fecal microbiome dysbiosis in patients with inflammatory bowel disease.Front Cell Infect Microbiol. 2022 Oct 7;12:1010853. doi: 10.3389/fcimb.2022.1010853. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36275026 Free PMC article.
-
Partners of patients with ulcerative colitis exhibit a biologically relevant dysbiosis in fecal microbial metacommunities.World J Gastroenterol. 2017 Jul 7;23(25):4624-4631. doi: 10.3748/wjg.v23.i25.4624. World J Gastroenterol. 2017. PMID: 28740351 Free PMC article.
-
Gut microbiota in mucosa and feces of newly diagnosed, treatment-naïve adult inflammatory bowel disease and irritable bowel syndrome patients.Gut Microbes. 2022 Jan-Dec;14(1):2083419. doi: 10.1080/19490976.2022.2083419. Gut Microbes. 2022. PMID: 35695669 Free PMC article.
-
Fecal transplantation for treatment of inflammatory bowel disease.Cochrane Database Syst Rev. 2018 Nov 13;11(11):CD012774. doi: 10.1002/14651858.CD012774.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2023 Apr 25;4:CD012774. doi: 10.1002/14651858.CD012774.pub3. PMID: 30480772 Free PMC article. Updated.
-
Fecal microbiota transplantation for ulcerative colitis.Immunol Med. 2021 Mar;44(1):30-34. doi: 10.1080/25785826.2020.1792040. Epub 2020 Jul 14. Immunol Med. 2021. PMID: 32663072 Review.
Cited by
-
The oral-gut microbiome axis in inflammatory bowel disease: from inside to insight.Front Immunol. 2024 Jul 26;15:1430001. doi: 10.3389/fimmu.2024.1430001. eCollection 2024. Front Immunol. 2024. PMID: 39131163 Free PMC article. Review.
-
Dental problems and oral microbiome alterations in ulcerative colitis.Front Immunol. 2025 Feb 5;16:1502605. doi: 10.3389/fimmu.2025.1502605. eCollection 2025. Front Immunol. 2025. PMID: 39975550 Free PMC article. Review.
-
Exploring non-invasive biomarkers for pulmonary nodule detection based on salivary microbiomics and machine learning algorithms.Sci Rep. 2025 Mar 29;15(1):10848. doi: 10.1038/s41598-025-95692-6. Sci Rep. 2025. PMID: 40155734 Free PMC article.
-
Neural network models for diagnosing recurrent aphthous ulcerations from clinical oral images.Sci Rep. 2025 Aug 12;15(1):29519. doi: 10.1038/s41598-025-06951-5. Sci Rep. 2025. PMID: 40796791 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- No. 7214267/the Beijing Municipal Natural Science Foundation
- No. 7232196/the Beijing Municipal Natural Science Foundation
- No. 82000496/the National Natural Science Foundation of China
- No. 82070539/the National Natural Science Foundation of China
- No. RDY2020-21/Peking University People's Hospital Scientific Research Development Funds
LinkOut - more resources
Full Text Sources
Medical

