YAP Signaling Regulates the Cellular Uptake and Therapeutic Effect of Nanoparticles
- PMID: 37946710
- PMCID: PMC10787066
- DOI: 10.1002/advs.202302965
YAP Signaling Regulates the Cellular Uptake and Therapeutic Effect of Nanoparticles
Abstract
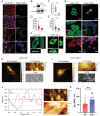
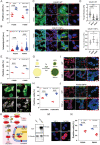
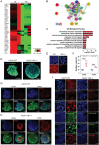
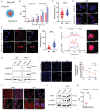
Interactions between living cells and nanoparticles are extensively studied to enhance the delivery of therapeutics. Nanoparticles size, shape, stiffness, and surface charge are regarded as the main features able to control the fate of cell-nanoparticle interactions. However, the clinical translation of nanotherapies has so far been limited, and there is a need to better understand the biology of cell-nanoparticle interactions. This study investigates the role of cellular mechanosensitive components in cell-nanoparticle interactions. It is demonstrated that the genetic and pharmacologic inhibition of yes-associated protein (YAP), a key component of cancer cell mechanosensing apparatus and Hippo pathway effector, improves nanoparticle internalization in triple-negative breast cancer cells regardless of nanoparticle properties or substrate characteristics. This process occurs through YAP-dependent regulation of endocytic pathways, cell mechanics, and membrane organization. Hence, the study proposes targeting YAP may sensitize triple-negative breast cancer cells to chemotherapy and increase the selectivity of nanotherapy.
Keywords: YAP-signaling; bio-nano interactions; cancer treatment; mechanobiology; nanoparticles.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S., Nature 2011, 474, 179. - PubMed
-
- Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R., Curr. Biol. 2007, 17, 2054. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NU23J-08-00035/Ministerstvo Zdravotnictví Ceské Republiky
- 800924/H2020 Marie Skłodowska-Curie Actions
- CZ.02.1.01/0.0/0.0/18_046/0015974/European Regional Development Fund
- CZ.02.1.01/0.0/0.0/16_019/0000868/European Regional Development Fund
- CZ.02.1.01/0.0/0.0/15_003/0000492/European Regional Development Fund
LinkOut - more resources
Full Text Sources
