Mechanisms Underlying Rare Inherited Pediatric Retinal Vascular Diseases: FEVR, Norrie Disease, Persistent Fetal Vascular Syndrome
- PMID: 37947657
- PMCID: PMC10647367
- DOI: 10.3390/cells12212579
Mechanisms Underlying Rare Inherited Pediatric Retinal Vascular Diseases: FEVR, Norrie Disease, Persistent Fetal Vascular Syndrome
Abstract
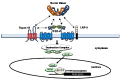
Familial Exudative Vitreoretinopathy (FEVR), Norrie disease, and persistent fetal vascular syndrome (PFVS) are extremely rare retinopathies that are clinically distinct but are unified by abnormal retinal endothelial cell function, and subsequent irregular retinal vascular development and/or aberrant inner blood-retinal-barrier (iBRB) function. The early angiogenesis of the retina and its iBRB is a delicate process that is mediated by the canonical Norrin Wnt-signaling pathway in retinal endothelial cells. Pathogenic variants in genes that play key roles within this pathway, such as NDP, FZD4, TSPAN12, and LRP5, have been associated with the incidence of these retinal diseases. Recent efforts to further elucidate the etiology of these conditions have not only highlighted their multigenic nature but have also resulted in the discovery of pathological variants in additional genes such as CTNNB1, KIF11, and ZNF408, some of which operate outside of the Norrin Wnt-signaling pathway. Recent discoveries of FEVR-linked variants in two other Catenin genes (CTNND1, CTNNA1) and the Endoplasmic Reticulum Membrane Complex Subunit-1 gene (EMC1) suggest that we will continue to find additional genes that impact the neural retinal vasculature, especially in multi-syndromic conditions. The goal of this review is to briefly highlight the current understanding of the roles of their encoded proteins in retinal endothelial cells to understand the essential functional mechanisms that can be altered to cause these very rare pediatric retinal vascular diseases.
Keywords: CTNNA1; CTNND1; ECM1; FEVR; FZD4; KIF11; LRP5; NDP; Norrie disease; TSPAN12; ZNF408; blood-brain-barrier; genetic disease mechanisms; norrin; persistent fetal vascular syndrome; retinal endothelial cell; retinal vasculature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Fruttiger M. Development of the Mouse Retinal Vasculature: Angiogenesis versus Vasculogenesis. Investig. Ophthalmol. Vis. Sci. 2002;43:522–527. - PubMed
-
- Tokunaga C.C., Mitton K.P., Dailey W., Massoll C., Roumayah K., Guzman E., Tarabishy N., Cheng M., Drenser K.A. Effects of Anti-VEGF Treatment on the Recovery of the Developing Retina Following Oxygen-Induced Retinopathy. Investig. Ophthalmol. Vis. Sci. 2014;55:1884–1892. doi: 10.1167/iovs.13-13397. - DOI - PubMed
-
- Dailey W.A., Drenser K.A., Wong S.C., Cheng M., Vercellone J., Roumayah K.K., Feeney E.V., Deshpande M., Guzman A.E., Trese M., et al. Ocular Coherence Tomography Image Data of the Retinal Laminar Structure in a Mouse Model of Oxygen-Induced Retinopathy. Data Brief. 2017;15:491–495. doi: 10.1016/j.dib.2017.09.075. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

