Deep phenotyping of the lipidomic response in COVID-19 and non-COVID-19 sepsis
- PMID: 37948331
- PMCID: PMC10637636
- DOI: 10.1002/ctm2.1440
Deep phenotyping of the lipidomic response in COVID-19 and non-COVID-19 sepsis
Abstract
Background: Lipids may influence cellular penetrance by viral pathogens and the immune response that they evoke. We deeply phenotyped the lipidomic response to SARs-CoV-2 and compared that with infection with other pathogens in patients admitted with acute respiratory distress syndrome to an intensive care unit (ICU).
Methods: Mass spectrometry was used to characterise lipids and relate them to proteins, peripheral cell immunotypes and disease severity.
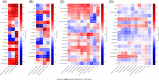
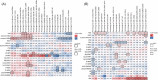
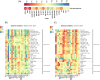
Results: Circulating phospholipases (sPLA2, cPLA2 (PLA2G4A) and PLA2G2D) were elevated on admission in all ICU groups. Cyclooxygenase, lipoxygenase and epoxygenase products of arachidonic acid (AA) were elevated in all ICU groups compared with controls. sPLA2 predicted severity in COVID-19 and correlated with TxA2, LTE4 and the isoprostane, iPF2α-III, while PLA2G2D correlated with LTE4. The elevation in PGD2, like PGI2 and 12-HETE, exhibited relative specificity for COVID-19 and correlated with sPLA2 and the interleukin-13 receptor to drive lymphopenia, a marker of disease severity. Pro-inflammatory eicosanoids remained correlated with severity in COVID-19 28 days after admission. Amongst non-COVID ICU patients, elevations in 5- and 15-HETE and 9- and 13-HODE reflected viral rather than bacterial disease. Linoleic acid (LA) binds directly to SARS-CoV-2 and both LA and its di-HOME products reflected disease severity in COVID-19. In healthy marines, these lipids rose with seroconversion. Eicosanoids linked variably to the peripheral cellular immune response. PGE2, TxA2 and LTE4 correlated with T cell activation, as did PGD2 with non-B non-T cell activation. In COVID-19, LPS stimulated peripheral blood mononuclear cell PGF2α correlated with memory T cells, dendritic and NK cells while LA and DiHOMEs correlated with exhausted T cells. Three high abundance lipids - ChoE 18:3, LPC-O-16:0 and PC-O-30:0 - were altered specifically in COVID. LPC-O-16:0 was strongly correlated with T helper follicular cell activation and all three negatively correlated with multi-omic inflammatory pathways and disease severity.
Conclusions: A broad based lipidomic storm is a predictor of poor prognosis in ARDS. Alterations in sPLA2, PGD2 and 12-HETE and the high abundance lipids, ChoE 18:3, LPC-O-16:0 and PC-O-30:0 exhibit relative specificity for COVID-19 amongst such patients and correlate with the inflammatory response to link to disease severity.
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
N. J. M. reports funding to her institution for unrelated work from Quantum Leap Healthcare Collaborative and BioMarck, Inc, and consulting fees from AstraZeneca Inc and Endpoint Health Inc. G. A. F. is the McNeil Professor of Translational Medicine and Therapeutics and held a Merit Award from the American Heart Association. He is a senior advisor to Calico Laboratories and serves on the scientific advisory boards of Bicycle Therapeutics and Kira Pharmaceuticals. A. G. L. is a military service member. This work was prepared as part of his official duties. Title 17, US Code §105 provides that copyright protection under this title is not available for any work of the US Government. Title 17, US code §101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person's official duties. The views expressed in the article are those of the authors and do not necessarily express the official policy and position of the US Navy, the Department of Defense, the US Government or the institutions affiliated with the authors. S. C. S. serves as Chief Scientific Officer for and holds equity in GNOMX Corp and has been a paid speaker for Illumina.
Figures




Update of
-
Deep Phenotyping of the Lipidomic Response in COVID and non-COVID Sepsis.bioRxiv [Preprint]. 2023 Jun 2:2023.06.02.543298. doi: 10.1101/2023.06.02.543298. bioRxiv. 2023. Update in: Clin Transl Med. 2023 Nov;13(11):e1440. doi: 10.1002/ctm2.1440. PMID: 37398323 Free PMC article. Updated. Preprint.
References
-
- Theken KN, FitzGerald GA. Bioactive lipids in antiviral immunity. Science. 2021;371(6526):237‐238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
