Antibody-mediated spike activation promotes cell-cell transmission of SARS-CoV-2
- PMID: 37948454
- PMCID: PMC10664894
- DOI: 10.1371/journal.ppat.1011789
Antibody-mediated spike activation promotes cell-cell transmission of SARS-CoV-2
Erratum in
-
Correction: Antibody-mediated spike activation promotes cell-cell transmission of SARS-CoV-2.PLoS Pathog. 2025 Aug 4;21(8):e1013394. doi: 10.1371/journal.ppat.1013394. eCollection 2025 Aug. PLoS Pathog. 2025. PMID: 40758617 Free PMC article.
Abstract
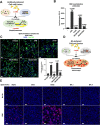
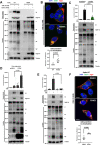
The COVID pandemic fueled by emerging SARS-CoV-2 new variants of concern remains a major global health concern, and the constantly emerging mutations present challenges to current therapeutics. The spike glycoprotein is not only essential for the initial viral entry, but is also responsible for the transmission of SARS-CoV-2 components via syncytia formation. Spike-mediated cell-cell transmission is strongly resistant to extracellular therapeutic and convalescent antibodies via an unknown mechanism. Here, we describe the antibody-mediated spike activation and syncytia formation on cells displaying the viral spike. We found that soluble antibodies against receptor binding motif (RBM) are capable of inducing the proteolytic processing of spike at both the S1/S2 and S2' cleavage sites, hence triggering ACE2-independent cell-cell fusion. Mechanistically, antibody-induced cell-cell fusion requires the shedding of S1 and exposure of the fusion peptide at the cell surface. By inhibiting S1/S2 proteolysis, we demonstrated that cell-cell fusion mediated by spike can be re-sensitized towards antibody neutralization in vitro. Lastly, we showed that cytopathic effect mediated by authentic SARS-CoV-2 infection remain unaffected by the addition of extracellular neutralization antibodies. Hence, these results unveil a novel mode of antibody evasion and provide insights for antibody selection and drug design strategies targeting the SARS-CoV-2 infected cells.
Copyright: © 2023 Yu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

