Transcriptomes and metabolism define mouse and human MAIT cell populations
- PMID: 37948512
- PMCID: PMC11160507
- DOI: 10.1126/sciimmunol.abn8531
Transcriptomes and metabolism define mouse and human MAIT cell populations
Erratum in
-
Erratum for the Research Article "Transcriptomes and metabolism define mouse and human MAIT cell populations" by S. Chandra et al.Sci Immunol. 2024 May 10;9(95):eadp9609. doi: 10.1126/sciimmunol.adp9609. Epub 2024 May 10. Sci Immunol. 2024. PMID: 38728415 No abstract available.
Abstract
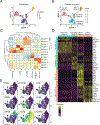
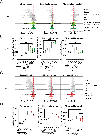
Mucosal-associated invariant T (MAIT) cells are a subset of T lymphocytes that respond to microbial metabolites. We defined MAIT cell populations in different organs and characterized the developmental pathway of mouse and human MAIT cells in the thymus using single-cell RNA sequencing and phenotypic and metabolic analyses. We showed that the predominant mouse subset, which produced IL-17 (MAIT17), and the subset that produced IFN-γ (MAIT1) had not only greatly different transcriptomes but also different metabolic states. MAIT17 cells in different organs exhibited increased lipid uptake, lipid storage, and mitochondrial potential compared with MAIT1 cells. All these properties were similar in the thymus and likely acquired there. Human MAIT cells in lung and blood were more homogeneous but still differed between tissues. Human MAIT cells had increased fatty acid uptake and lipid storage in blood and lung, similar to human CD8 T resident memory cells, but unlike mouse MAIT17 cells, they lacked increased mitochondrial potential. Although mouse and human MAIT cell transcriptomes showed similarities for immature cells in the thymus, they diverged more strikingly in the periphery. Analysis of pet store mice demonstrated decreased lung MAIT17 cells in these so-called "dirty" mice, indicative of an environmental influence on MAIT cell subsets and function.
Figures





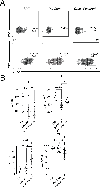
References
-
- Treiner E et al., Mucosal-associated invariant T (MAIT) cells: an evolutionarily conserved T cell subset. Microbes Infect 7, 552–559 (2005). - PubMed
-
- Kjer-Nielsen L et al., MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). - PubMed
-
- Treiner E et al., Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 OD016262/OD/NIH HHS/United States
- S10 OD025052/OD/NIH HHS/United States
- S10 RR027366/RR/NCRR NIH HHS/United States
- R01 AI105215/AI/NIAID NIH HHS/United States
- R01 AI137230/AI/NIAID NIH HHS/United States
- R37 AI071922/AI/NIAID NIH HHS/United States
- 210842/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI071922/AI/NIAID NIH HHS/United States
- R21 AI163813/AI/NIAID NIH HHS/United States
- 210842_Z_18_Z/WT_/Wellcome Trust/United Kingdom
- DH_/Department of Health/United Kingdom
- R01 AI108651/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

