Host Response Changes and Their Association with Mortality in COVID-19 Patients with Lymphopenia
- PMID: 37948687
- PMCID: PMC10878379
- DOI: 10.1164/rccm.202305-0890OC
Host Response Changes and Their Association with Mortality in COVID-19 Patients with Lymphopenia
Abstract
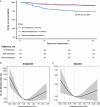
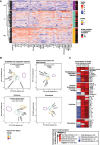
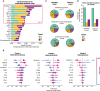
Rationale: Lymphopenia in coronavirus disease (COVID-19) is associated with increased mortality. Objectives: To explore the association between lymphopenia, host response aberrations, and mortality in patients with lymphopenic COVID-19. Methods: We determined 43 plasma biomarkers reflective of four pathophysiological domains: endothelial cell and coagulation activation, inflammation and organ damage, cytokine release, and chemokine release. We explored if decreased concentrations of lymphocyte-derived proteins in patients with lymphopenia were associated with an increase in mortality. We sought to identify host response phenotypes in patients with lymphopenia by cluster analysis of plasma biomarkers. Measurements and Main Results: A total of 439 general ward patients with COVID-19 were stratified by baseline lymphocyte counts: normal (>1.0 × 109/L; n = 167), mild lymphopenia (>0.5 to ⩽1.0 × 109/L; n = 194), and severe lymphopenia (⩽0.5 × 109/L; n = 78). Lymphopenia was associated with alterations in each host response domain. Lymphopenia was associated with increased mortality. Moreover, in patients with lymphopenia (n = 272), decreased concentrations of several lymphocyte-derived proteins (e.g., CCL5, IL-4, IL-13, IL-17A) were associated with an increase in mortality (at P < 0.01 or stronger significance levels). A cluster analysis revealed three host response phenotypes in patients with lymphopenia: "hyporesponsive" (23.2%), "hypercytokinemic" (36.4%), and "inflammatory-injurious" (40.4%), with substantially differing mortality rates of 9.5%, 5.1%, and 26.4%, respectively. A 10-biomarker model accurately predicted these host response phenotypes in an external cohort with similar mortality distribution. The inflammatory-injurious phenotype showed a remarkable combination of relatively high inflammation and organ damage markers with high antiinflammatory cytokine levels yet low proinflammatory cytokine levels. Conclusions: Lymphopenia in COVID-19 signifies a heterogenous group of patients with distinct host response features. Specific host responses contribute to lymphopenia-associated mortality in COVID-19, including reduced CCL5 levels.
Keywords: biomarkers; cytokines; lymphocytes; phenotype; pneumonia.
Figures



Comment in
-
Host Response to Infection: Not All Lymphopenia Is Created Equal in SARS-CoV-2.Am J Respir Crit Care Med. 2024 Feb 15;209(4):351-352. doi: 10.1164/rccm.202312-2265ED. Am J Respir Crit Care Med. 2024. PMID: 38190496 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

