Interaction between the mitochondrial adaptor MIRO and the motor adaptor TRAK
- PMID: 37949220
- PMCID: PMC10746525
- DOI: 10.1016/j.jbc.2023.105441
Interaction between the mitochondrial adaptor MIRO and the motor adaptor TRAK
Abstract
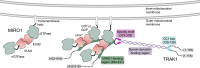
MIRO (mitochondrial Rho GTPase) consists of two GTPase domains flanking two Ca2+-binding EF-hand domains. A C-terminal transmembrane helix anchors MIRO to the outer mitochondrial membrane, where it functions as a general adaptor for the recruitment of cytoskeletal proteins that control mitochondrial dynamics. One protein recruited by MIRO is TRAK (trafficking kinesin-binding protein), which in turn recruits the microtubule-based motors kinesin-1 and dynein-dynactin. The mechanism by which MIRO interacts with TRAK is not well understood. Here, we map and quantitatively characterize the interaction of human MIRO1 and TRAK1 and test its potential regulation by Ca2+ and/or GTP binding. TRAK1 binds MIRO1 with low micromolar affinity. The interaction was mapped to a fragment comprising MIRO1's EF-hands and C-terminal GTPase domain and to a conserved sequence motif within TRAK1 residues 394 to 431, immediately C-terminal to the Spindly motif. This sequence is sufficient for MIRO1 binding in vitro and is necessary for MIRO1-dependent localization of TRAK1 to mitochondria in cells. MIRO1's EF-hands bind Ca2+ with dissociation constants (KD) of 3.9 μM and 300 nM. This suggests that under cellular conditions one EF-hand may be constitutively bound to Ca2+ whereas the other EF-hand binds Ca2+ in a regulated manner, depending on its local concentration. Yet, the MIRO1-TRAK1 interaction is independent of Ca2+ binding to the EF-hands and of the nucleotide state (GDP or GTP) of the C-terminal GTPase. The interaction is also independent of TRAK1 dimerization, such that a TRAK1 dimer can be expected to bind two MIRO1 molecules on the mitochondrial surface.
Keywords: EF-hand; GTPase; calcium; isothermal titration calorimetry (ITC); mitochondrial dynamics; motor adaptor; mutagenesis.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Fransson A., Ruusala A., Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003;278:6495–6502. - PubMed
-
- Fransson S., Ruusala A., Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 2006;344:500–510. - PubMed
-
- Birsa N., Norkett R., Higgs N., Lopez-Domenech G., Kittler J.T. Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Biochem. Soc. Trans. 2013;41:1525–1531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

