Near-Infrared Optogenetic Module for Conditional Protein Splicing
- PMID: 37949312
- PMCID: PMC10842711
- DOI: 10.1016/j.jmb.2023.168360
Near-Infrared Optogenetic Module for Conditional Protein Splicing
Abstract
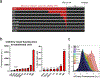
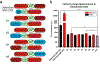
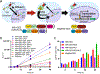
Optogenetics has emerged as a powerful tool for spatiotemporal control of biological processes. Near-infrared (NIR) light, with its low phototoxicity and deep tissue penetration, holds particular promise. However, the optogenetic control of polypeptide bond formation has not yet been developed. In this study, we introduce a NIR optogenetic module for conditional protein splicing (CPS) based on the gp41-1 intein. We optimized the module to minimize background signals in the darkness and to maximize the contrast between light and dark conditions. Next, we engineered a NIR CPS gene expression system based on the protein ligation of a transcription factor. We applied the NIR CPS for light-triggered protein cleavage to activate gasdermin D, a pore-forming protein that induces pyroptotic cell death. Our NIR CPS optogenetic module represents a promising tool for controlling molecular processes through covalent protein linkage and cleavage.
Keywords: DrBphP; MagRed; pyroptosis; split intein; tTA.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
An intein-cassette integration approach used for the generation of a split TEV protease activated by conditional protein splicing.Mol Biosyst. 2011 Jun;7(6):2031-9. doi: 10.1039/c1mb05025g. Epub 2011 Apr 12. Mol Biosyst. 2011. PMID: 21487580
-
Reactive Chlorine Species Reversibly Inhibit DnaB Protein Splicing in Mycobacteria.Microbiol Spectr. 2021 Oct 31;9(2):e0030121. doi: 10.1128/Spectrum.00301-21. Epub 2021 Sep 22. Microbiol Spectr. 2021. PMID: 34549994 Free PMC article.
-
Engineering artificially split inteins for applications in protein chemistry: biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein.Biochemistry. 2006 Feb 14;45(6):1571-8. doi: 10.1021/bi051697+. Biochemistry. 2006. PMID: 16460004
-
[Protein splicing and its application].Sheng Wu Gong Cheng Xue Bao. 2003 Mar;19(2):249-54. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15966332 Review. Chinese.
-
Near-Infrared-Light Activatable Nanoparticles for Deep-Tissue-Penetrating Wireless Optogenetics.Adv Healthc Mater. 2019 Mar;8(6):e1801132. doi: 10.1002/adhm.201801132. Epub 2019 Jan 11. Adv Healthc Mater. 2019. PMID: 30633858 Review.
Cited by
-
Optogenetic engineering for precision cancer immunotherapy.Trends Pharmacol Sci. 2025 Jun 10:S0165-6147(25)00092-6. doi: 10.1016/j.tips.2025.05.002. Online ahead of print. Trends Pharmacol Sci. 2025. PMID: 40500674 Review.
References
-
- Adam E, & Perler FB (2002). Development of a positive genetic selection system for inhibition of protein splicing using mycobacterial inteins in Escherichia coli DNA gyrase subunit A. Journal of Molecular Microbiology and Biotechnology, 4(5), 479–487. - PubMed
-
- Beyer HM, Juillot S, Herbst K, Samodelov SL, Müller K, Schamel WW, Römer W, Schäfer E, Nagy F, Strähle U, Weber W, & Zurbriggen MD (2015). Red Light-Regulated Reversible Nuclear Localization of Proteins in Mammalian Cells and Zebrafish. ACS Synthetic Biology, 4(9), 951–958. 10.1021/acssynbio.5b00004 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

