Visualization of conformational transition of GRP94 in solution
- PMID: 37949474
- PMCID: PMC10638095
- DOI: 10.26508/lsa.202302051
Visualization of conformational transition of GRP94 in solution
Abstract
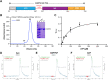
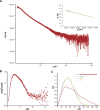
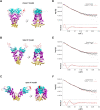
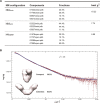
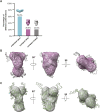
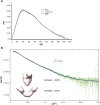
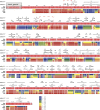
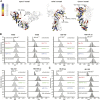
GRP94, an ER paralog of the heat-shock protein 90 family, binds and hydrolyses ATP to chaperone the folding and maturation of its selected clients. Compared with other hsp90 proteins, the in-solution conformational dynamics of GRP94 along the ATP hydrolysis cycle are less understood, hindering our understanding of its chaperoning mechanism. Leveraging small-angle X-ray scattering, negative-staining EM, and hydrogen-deuterium exchange coupled mass-spec, here we show that in its apo form, ∼60% of mouse GRP94 (mGRP94) populates an "extended" conformation, whereas the rest exist in either "close V" or "twist V" like "compact" conformations. Different from other hsp90 proteins, the presence of AMPPNP only impacts the relative abundance of the two compact conformations, rather than shifting the equilibrium between the "extended" and "compact" conformations of mGRP94. HDX-MS study of apo, AMPPNP-bound, and ADP-bound mGRP94 suggests a conformational transition from "twist V" to "close V" upon ATP binding and a back transition from "close V" to "twist V" upon ATP hydrolysis. These results illustrate the dissimilarities of GRP94 in conformation transition during ATP hydrolysis from other hsp90 paralogs.
© 2023 Sun et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
