Single-Cell Analysis Reveals Malignant Cells Reshape the Cellular Landscape and Foster an Immunosuppressive Microenvironment of Extranodal NK/T-Cell Lymphoma
- PMID: 37949673
- PMCID: PMC10754138
- DOI: 10.1002/advs.202303913
Single-Cell Analysis Reveals Malignant Cells Reshape the Cellular Landscape and Foster an Immunosuppressive Microenvironment of Extranodal NK/T-Cell Lymphoma
Abstract
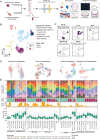
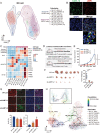
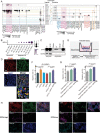
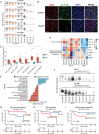
Extranodal natural killer/T-cell lymphoma (NKTCL) is an aggressive type of lymphoma associated with Epstein-Barr virus (EBV) and characterized by heterogeneous tumor behaviors. To better understand the origins of the heterogeneity, this study utilizes single-cell RNA sequencing (scRNA-seq) analysis to profile the tumor microenvironment (TME) of NKTCL at the single-cell level. Together with in vitro and in vivo models, the study identifies a subset of LMP1+ malignant NK cells contributing to the tumorigenesis and development of heterogeneous malignant cells in NKTCL. Furthermore, malignant NK cells interact with various immunocytes via chemokines and their receptors, secrete substantial DPP4 that impairs the chemotaxis of immunocytes and regulates their infiltration. They also exhibit an immunosuppressive effect on T cells, which is further boosted by LMP1. Moreover, high transcription of EBV-encoded genes and low infiltration of tumor-associated macrophages (TAMs) are favorable prognostic indicators for NKTCL in multiple patient cohorts. This study for the first time deciphers the heterogeneous composition of NKTCL TME at single-cell resolution, highlighting the crucial role of malignant NK cells with EBV-encoded LMP1 in reshaping the cellular landscape and fostering an immunosuppressive microenvironment. These findings provide insights into understanding the pathogenic mechanisms of NKTCL and developing novel therapeutic strategies against NKTCL.
Keywords: DPP4; LMP1+ malignant NK cells; NKTCL; immunosuppression; scRNA-seq.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



References
-
- Huang H., Gao Y., Wang X., Cai Q., Cai Q., Bai B., Zhao W., Yan Z., Jiang W., Xia Z., Li Z., Blood 2015, 126, 1521.
-
- Yamaguchi M., Tobinai K., Oguchi M., Ishizuka N., Kobayashi Y., Isobe Y., Ishizawa K., Maseki N., Itoh K., Usui N., Wasada I., Kinoshita T., Ohshima K., Matsuno Y., Terauchi T., Nawano S., Ishikura S., Kagami Y., Hotta T., Oshimi K., J. Clin. Oncol. 2009, 27, 5594. - PubMed
-
- Kim S. J., Yang D.‐H., Kim J. S., Kwak J.‐Y., Eom H.‐S., Hong D. S., Won J Ho, Lee J. H., Yoon D. H., Cho J., Nam T.‐K., Lee S.‐W., Ahn Y. C., Suh C., Kim W. S., Ann. Hematol. 2014, 93, 1895. - PubMed
-
- Yamaguchi M., Kwong Y.‐L., Kim W. S., Maeda Y., Hashimoto C., Suh C., Izutsu K., Ishida F., Isobe Y., Sueoka E., Suzumiya J., Kodama T., Kimura H., Hyo R., Nakamura S., Oshimi K., Suzuki R., J. Clin. Oncol. 2011, 29, 4410. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
