A large meta-analysis identifies genes associated with anterior uveitis
- PMID: 37949852
- PMCID: PMC10638276
- DOI: 10.1038/s41467-023-43036-1
A large meta-analysis identifies genes associated with anterior uveitis
Abstract
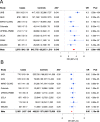
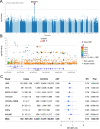
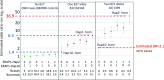
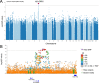
Anterior Uveitis (AU) is the inflammation of the anterior part of the eye, the iris and ciliary body and is strongly associated with HLA-B*27. We report AU exome sequencing results from eight independent cohorts consisting of 3,850 cases and 916,549 controls. We identify common genome-wide significant loci in HLA-B (OR = 3.37, p = 1.03e-196) and ERAP1 (OR = 0.86, p = 1.1e-08), and find IPMK (OR = 9.4, p = 4.42e-09) and IDO2 (OR = 3.61, p = 6.16e-08) as genome-wide significant genes based on the burden of rare coding variants. Dividing the cohort into HLA-B*27 positive and negative individuals, we find ERAP1 haplotype is strongly protective only for B*27-positive AU (OR = 0.73, p = 5.2e-10). Investigation of B*27-negative AU identifies a common signal near HLA-DPB1 (rs3117230, OR = 1.26, p = 2.7e-08), risk genes IPMK and IDO2, and several additional candidate risk genes, including ADGFR5, STXBP2, and ACHE. Taken together, we decipher the genetics underlying B*27-positive and -negative AU and identify rare and common genetic signals for both subtypes of disease.
© 2023. The Author(s).
Conflict of interest statement
S.G., A.M., S.M.H., R.W., V.R., N.P., V.K.P., E.C., M.L., R.H., J.F., B.C., A.J.L., C.G.M., T.V., J.W., C.R., B.S., A.B., E.A.S. and G.C. are current employees and/or stockholders of Regeneron Genetics Center or Regeneron Pharmaceuticals. All other authors declare no conflict of interest.
Figures

References
-
- Rosenbaum JT, et al. New observations and emerging ideas in diagnosis and management of non-infectious uveitis: A review. Semin Arthritis Rheum. 2019;49:438–445. - PubMed
-
- Thorne JE, et al. Prevalence of noninfectious uveitis in the United States: A claims-based analysis. JAMA Ophthalmol. 2016;134:1237–1245. - PubMed
-
- Chang JH, Wakefield D. Uveitis: a global perspective. Ocul. Immunol. Inflamm. 2002;10:263–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

