A pathway activity-based proteomic classifier stratifies prostate tumors into two subtypes
- PMID: 37950160
- PMCID: PMC10638831
- DOI: 10.1186/s12014-023-09441-w
A pathway activity-based proteomic classifier stratifies prostate tumors into two subtypes
Abstract
Prostate cancer (PCa) is the second most common cancer in males worldwide. The risk stratification of PCa is mainly based on morphological examination. Here we analyzed the proteome of 667 tumor samples from 487 Chinese PCa patients and characterized 9576 protein groups by PulseDIA mass spectrometry. Then we developed a pathway activity-based classifier concerning 13 proteins from seven pathways, and dichotomized the PCa patients into two subtypes, namely PPS1 and PPS2. PPS1 is featured with enhanced innate immunity, while PPS2 with suppressed innate immunity. This classifier exhibited a correlation with PCa progression in our cohort and was further validated by two published transcriptome datasets. Notably, PPS2 was significantly correlated with poor biochemical recurrence (BCR)/metastasis-free survival (log-rank P-value < 0.05). The PPS2 was also featured with cell proliferation activation. Together, our study presents a novel pathway activity-based stratification scheme for PCa.
Keywords: BCR-free survival; Prostate cancer; Proteomic pathway-based classifier; PulseDIA.
© 2023. The Author(s).
Conflict of interest statement
T.G. is a shareholder of Westlake Omics Inc. L.T. is an employee of Westlake Omics Inc. The other authors declare no competing interests in this paper.
Figures

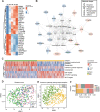
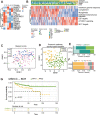

References
-
- Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–52. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
