Role of environmental specificity in CASP results
- PMID: 37950210
- PMCID: PMC10638730
- DOI: 10.1186/s12859-023-05559-8
Role of environmental specificity in CASP results
Abstract
Background: Recently, significant progress has been made in the field of protein structure prediction by the application of artificial intelligence techniques, as shown by the results of the CASP13 and CASP14 (Critical Assessment of Structure Prediction) competition. However, the question of the mechanism behind the protein folding process itself remains unanswered. Correctly predicting the structure also does not solve the problem of, for example, amyloid proteins, where a polypeptide chain with an unaltered sequence adopts a different 3D structure.
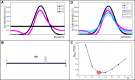
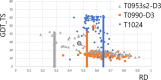
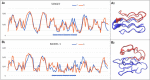
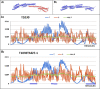
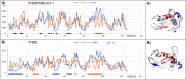
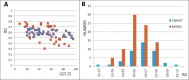
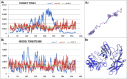
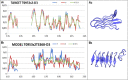
Results: This work was an attempt at explaining the structural variation by considering the contribution of the environment to protein structuring. The application of the fuzzy oil drop (FOD) model to assess the validity of the selected models provided in the CASP13, CASP14 and CASP15 projects reveals the need for an environmental factor to determine the 3D structure of proteins. Consideration of the external force field in the form of polar water (Fuzzy Oil Drop) and a version modified by the presence of the hydrophobic compounds, FOD-M (FOD-Modified) reveals that the protein folding process is environmentally dependent. An analysis of selected models from the CASP competitions indicates the need for structure prediction as dependent on the consideration of the protein folding environment.
Conclusions: The conditions governed by the environment direct the protein folding process occurring in a certain environment. Therefore, the variation of the external force field should be taken into account in the models used in protein structure prediction.
Keywords: CASP; Folding environment; Folding simulation in Silico; Protein folding.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- https://predictioncenter.org/ (accessed Aug 7, 2023)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

