Bcl6 is a subset-defining transcription factor of lymphoid tissue inducer-like ILC3
- PMID: 37950867
- PMCID: PMC7615641
- DOI: 10.1016/j.celrep.2023.113425
Bcl6 is a subset-defining transcription factor of lymphoid tissue inducer-like ILC3
Abstract
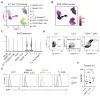
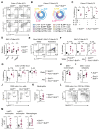
Innate lymphoid cells (ILCs) are tissue-resident effector cells with roles in tissue homeostasis, protective immunity, and inflammatory disease. Group 3 ILCs (ILC3s) are classically defined by the master transcription factor RORγt. However, ILC3 can be further subdivided into subsets that share type 3 effector modules that exhibit significant ontological, transcriptional, phenotypic, and functional heterogeneity. Notably lymphoid tissue inducer (LTi)-like ILC3s mediate effector functions not typically associated with other RORγt-expressing lymphocytes, suggesting that additional transcription factors contribute to dictate ILC3 subset phenotypes. Here, we identify Bcl6 as a subset-defining transcription factor of LTi-like ILC3s in mice and humans. Deletion of Bcl6 results in dysregulation of the LTi-like ILC3 transcriptional program and markedly enhances expression of interleukin-17A (IL-17A) and IL-17F in LTi-like ILC3s in a manner in part dependent upon the commensal microbiota-and associated with worsened inflammation in a model of colitis. Together, these findings redefine our understanding of ILC3 subset biology.
Keywords: Bcl6; CP: Immunology; IL-17; ILC3; innate lymphoid cells; intestinal immunity; lymphoid tissue inducer; microbiota; transcription factor.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

