Anti-tumor activity of all-trans retinoic acid in gastric-cancer: gene-networks and molecular mechanisms
- PMID: 37951921
- PMCID: PMC10638833
- DOI: 10.1186/s13046-023-02869-w
Anti-tumor activity of all-trans retinoic acid in gastric-cancer: gene-networks and molecular mechanisms
Abstract
Background: Gastric-cancer is a heterogeneous type of neoplastic disease and it lacks appropriate therapeutic options. There is an urgent need for the development of innovative pharmacological strategies, particularly in consideration of the potential stratified/personalized treatment of this tumor. All-Trans Retinoic-acid (ATRA) is one of the active metabolites of vitamin-A. This natural compound is the first example of clinically approved cyto-differentiating agent, being used in the treatment of acute promyelocytic leukemia. ATRA may have significant therapeutic potential also in the context of solid tumors, including gastric-cancer. The present study provides pre-clinical evidence supporting the use of ATRA in the treatment of gastric-cancer using high-throughput approaches.
Methods: We evaluated the anti-proliferative action of ATRA in 27 gastric-cancer cell-lines and tissue-slice cultures from 13 gastric-cancer patients. We performed RNA-sequencing studies in 13 cell-lines exposed to ATRA. We used these and the gastric-cancer RNA-sequencing data of the TCGA/CCLE datasets to conduct multiple computational analyses.
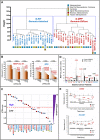
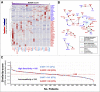
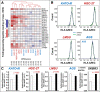
Results: Profiling of our large panel of gastric-cancer cell-lines for their quantitative response to the anti-proliferative effects of ATRA indicate that approximately half of the cell-lines are characterized by sensitivity to the retinoid. The constitutive transcriptomic profiles of these cell-lines permitted the construction of a model consisting of 42 genes, whose expression correlates with ATRA-sensitivity. The model predicts that 45% of the TCGA gastric-cancers are sensitive to ATRA. RNA-sequencing studies performed in retinoid-treated gastric-cancer cell-lines provide insights into the gene-networks underlying ATRA anti-tumor activity. In addition, our data demonstrate that ATRA exerts significant immune-modulatory effects, which seem to be largely controlled by IRF1 up-regulation. Finally, we provide evidence of a feed-back loop between IRF1 and DHRS3, another gene which is up-regulated by ATRA.
Conclusions: ATRA is endowed with significant therapeutic potential in the stratified/personalized treatment gastric-cancer. Our data represent the fundaments for the design of clinical trials focusing on the use of ATRA in the personalized treatment of this heterogeneous tumor. Our gene-expression model will permit the development of a predictive tool for the selection of ATRA-sensitive gastric-cancer patients. The immune-regulatory responses activated by ATRA suggest that the retinoid and immune-checkpoint inhibitors constitute rational combinations for the management of gastric-cancer.
Keywords: All-trans-retinoic-acid; Gastric-cancer; IRF1; Immune-responses; RNA-sequencing.
© 2023. The Author(s).
Conflict of interest statement
All the authors declare that they have no conflict of interest as to the content of the present manuscript.
Figures





Similar articles
-
Cellular and molecular determinants of all-trans retinoic acid sensitivity in breast cancer: Luminal phenotype and RARα expression.EMBO Mol Med. 2015 Jul;7(7):950-72. doi: 10.15252/emmm.201404670. EMBO Mol Med. 2015. PMID: 25888236 Free PMC article.
-
Network-guided modeling allows tumor-type independent prediction of sensitivity to all-trans-retinoic acid.Ann Oncol. 2017 Mar 1;28(3):611-621. doi: 10.1093/annonc/mdw660. Ann Oncol. 2017. PMID: 27993792 Free PMC article.
-
All-Trans Retinoic Acid Stimulates Viral Mimicry, Interferon Responses and Antigen Presentation in Breast-Cancer Cells.Cancers (Basel). 2020 May 6;12(5):1169. doi: 10.3390/cancers12051169. Cancers (Basel). 2020. PMID: 32384653 Free PMC article.
-
Efficiency of All-Trans Retinoic Acid on Gastric Cancer: A Narrative Literature Review.Int J Mol Sci. 2018 Oct 29;19(11):3388. doi: 10.3390/ijms19113388. Int J Mol Sci. 2018. PMID: 30380687 Free PMC article. Review.
-
Cellular and micro-environmental responses influencing the antitumor activity of all-trans retinoic acid in breast cancer.Cell Commun Signal. 2024 Feb 15;22(1):127. doi: 10.1186/s12964-024-01492-2. Cell Commun Signal. 2024. PMID: 38360674 Free PMC article. Review.
Cited by
-
Preclinical evaluation of fenretinide against primary and metastatic intestinal type‑gastric cancer.Oncol Lett. 2024 Sep 25;28(6):561. doi: 10.3892/ol.2024.14694. eCollection 2024 Dec. Oncol Lett. 2024. PMID: 39372665 Free PMC article.
-
Fuelling the Fight from the Gut: Short-Chain Fatty Acids and Dexamethasone Synergise to Suppress Gastric Cancer Cells.Cancers (Basel). 2025 Jul 28;17(15):2486. doi: 10.3390/cancers17152486. Cancers (Basel). 2025. PMID: 40805185 Free PMC article.
-
Identification of prognostic biomarkers related to retinoic acid metabolism in gliomas and analysis of their impact on the immune microenvironment.Medicine (Baltimore). 2024 Oct 11;103(41):e39836. doi: 10.1097/MD.0000000000039836. Medicine (Baltimore). 2024. PMID: 39465792 Free PMC article.
-
All-trans retinoic acid in hematologic disorders: not just acute promyelocytic leukemia.Front Pharmacol. 2024 Jul 4;15:1404092. doi: 10.3389/fphar.2024.1404092. eCollection 2024. Front Pharmacol. 2024. PMID: 39027338 Free PMC article. Review.
-
Path-based reasoning for biomedical knowledge graphs with BioPathNet.bioRxiv [Preprint]. 2024 Aug 10:2024.06.17.599219. doi: 10.1101/2024.06.17.599219. bioRxiv. 2024. PMID: 39149355 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

