The DO-KB Knowledgebase: a 20-year journey developing the disease open science ecosystem
- PMID: 37953304
- PMCID: PMC10767934
- DOI: 10.1093/nar/gkad1051
The DO-KB Knowledgebase: a 20-year journey developing the disease open science ecosystem
Abstract
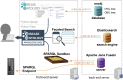
In 2003, the Human Disease Ontology (DO, https://disease-ontology.org/) was established at Northwestern University. In the intervening 20 years, the DO has expanded to become a highly-utilized disease knowledge resource. Serving as the nomenclature and classification standard for human diseases, the DO provides a stable, etiology-based structure integrating mechanistic drivers of human disease. Over the past two decades the DO has grown from a collection of clinical vocabularies, into an expertly curated semantic resource of over 11300 common and rare diseases linking disease concepts through more than 37000 vocabulary cross mappings (v2023-08-08). Here, we introduce the recently launched DO Knowledgebase (DO-KB), which expands the DO's representation of the diseaseome and enhances the findability, accessibility, interoperability and reusability (FAIR) of disease data through a new SPARQL service and new Faceted Search Interface. The DO-KB is an integrated data system, built upon the DO's semantic disease knowledge backbone, with resources that expose and connect the DO's semantic knowledge with disease-related data across Open Linked Data resources. This update includes descriptions of efforts to assess the DO's global impact and improvements to data quality and content, with emphasis on changes in the last two years.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

