A Cre-dependent massively parallel reporter assay allows for cell-type specific assessment of the functional effects of non-coding elements in vivo
- PMID: 37953348
- PMCID: PMC10641075
- DOI: 10.1038/s42003-023-05483-w
A Cre-dependent massively parallel reporter assay allows for cell-type specific assessment of the functional effects of non-coding elements in vivo
Abstract
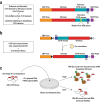
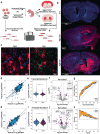
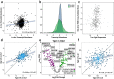
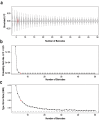
The function of regulatory elements is highly dependent on the cellular context, and thus for understanding the function of elements associated with psychiatric diseases these would ideally be studied in neurons in a living brain. Massively Parallel Reporter Assays (MPRAs) are molecular genetic tools that enable functional screening of hundreds of predefined sequences in a single experiment. These assays have not yet been adapted to query specific cell types in vivo in a complex tissue like the mouse brain. Here, using a test-case 3'UTR MPRA library with genomic elements containing variants from autism patients, we developed a method to achieve reproducible measurements of element effects in vivo in a cell type-specific manner, using excitatory cortical neurons and striatal medium spiny neurons as test cases. This targeted technique should enable robust, functional annotation of genetic elements in the cellular contexts most relevant to psychiatric disease.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hevner RF, Hodge RD, Daza RAM, Englund C. Transcription factors in glutamatergic neurogenesis: Conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci. Res. 2006;55:223–233. - PubMed

