Vesiclepedia 2024: an extracellular vesicles and extracellular particles repository
- PMID: 37953359
- PMCID: PMC10767981
- DOI: 10.1093/nar/gkad1007
Vesiclepedia 2024: an extracellular vesicles and extracellular particles repository
Abstract
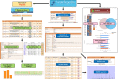
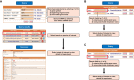
Vesiclepedia (http://www.microvesicles.org) is a free web-based compendium of DNA, RNA, proteins, lipids and metabolites that are detected or associated with extracellular vesicles (EVs) and extracellular particles (EPs). EVs are membranous vesicles that are secreted ubiquitously by cells from all domains of life from archaea to eukaryotes. In addition to EVs, it was reported recently that EPs like exomeres and supermeres are secreted by some mammalian cells. Both EVs and EPs contain proteins, nucleic acids, lipids and metabolites and has been proposed to be implicated in several key biological functions. Vesiclepedia catalogues proteins, DNA, RNA, lipids and metabolites from both published and unpublished studies. Currently, Vesiclepedia contains data obtained from 3533 EV studies, 50 550 RNA entries, 566 911 protein entries, 3839 lipid entries, 192 metabolite and 167 DNA entries. Quantitative data for 62 822 entries from 47 EV studies is available in Vesiclepedia. The datasets available in Vesiclepedia can be downloaded as tab-delimited files or accessible through the FunRich-based Vesiclepedia plugin.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Zhang Q., Jeppesen D.K., Higginbotham J.N., Graves-Deal R., Trinh V.Q., Ramirez M.A., Sohn Y., Neininger A.C., Taneja N., McKinley E.T.et al. .. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021; 23:1240–1254. - PMC - PubMed
-
- Anand S., Samuel M., Mathivanan S.. Exomeres: a new member of extracellular vesicles family. Subcell. Biochem. 2021; 97:89–97. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

