DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms
- PMID: 37953380
- PMCID: PMC10767982
- DOI: 10.1093/nar/gkad1040
DGIdb 5.0: rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms
Abstract
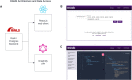
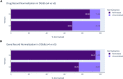
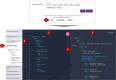
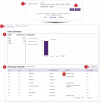
The Drug-Gene Interaction Database (DGIdb, https://dgidb.org) is a publicly accessible resource that aggregates genes or gene products, drugs and drug-gene interaction records to drive hypothesis generation and discovery for clinicians and researchers. DGIdb 5.0 is the latest release and includes substantial architectural and functional updates to support integration into clinical and drug discovery pipelines. The DGIdb service architecture has been split into separate client and server applications, enabling consistent data access for users of both the application programming interface (API) and web interface. The new interface was developed in ReactJS, and includes dynamic visualizations and consistency in the display of user interface elements. A GraphQL API has been added to support customizable queries for all drugs, genes, annotations and associated data. Updated documentation provides users with example queries and detailed usage instructions for these new features. In addition, six sources have been added and many existing sources have been updated. Newly added sources include ChemIDplus, HemOnc, NCIt (National Cancer Institute Thesaurus), Drugs@FDA, HGNC (HUGO Gene Nomenclature Committee) and RxNorm. These new sources have been incorporated into DGIdb to provide additional records and enhance annotations of regulatory approval status for therapeutics. Methods for grouping drugs and genes have been expanded upon and developed as independent modular normalizers during import. The updates to these sources and grouping methods have resulted in an improvement in FAIR (findability, accessibility, interoperability and reusability) data representation in DGIdb.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

