The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered
- PMID: 37953409
- PMCID: PMC10767974
- DOI: 10.1093/nar/gkad1039
The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered
Abstract
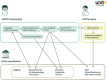
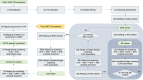
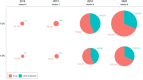
UNITE (https://unite.ut.ee) is a web-based database and sequence management environment for molecular identification of eukaryotes. It targets the nuclear ribosomal internal transcribed spacer (ITS) region and offers nearly 10 million such sequences for reference. These are clustered into ∼2.4M species hypotheses (SHs), each assigned a unique digital object identifier (DOI) to promote unambiguous referencing across studies. UNITE users have contributed over 600 000 third-party sequence annotations, which are shared with a range of databases and other community resources. Recent improvements facilitate the detection of cross-kingdom biological associations and the integration of undescribed groups of organisms into everyday biological pursuits. Serving as a digital twin for eukaryotic biodiversity and communities worldwide, the latest release of UNITE offers improved avenues for biodiversity discovery, precise taxonomic communication and integration of biological knowledge across platforms.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W., Bolchacova E., Voigt K., Crous P.W.et al.. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6241–6246. - PMC - PubMed
-
- Taberlet P., Coissac E., Pompanon F., Brochmann C., Willerslew E.. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012; 21:2045–2050. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

