Different gut microbial types were found in captive striped hamsters
- PMID: 37953783
- PMCID: PMC10634337
- DOI: 10.7717/peerj.16365
Different gut microbial types were found in captive striped hamsters
Abstract
Background: Typing analysis has become a popular approach to categorize individual differences in studies of animal gut microbial communities. However, previous definitions of gut microbial types were more understood as a passive reaction process to different external interferences, as most studies involve diverse environmental variables. We wondered whether distinct gut microbial types can also occur in animals under the same external environment. Moreover, the role of host sex in shaping gut microbiota has been widely reported; thus, the current study preliminarily explores the effects of sex on potential different microbial types.
Methods: Here, adult striped hamsters Cricetulus barabensis of different sexes were housed under the same controlled laboratory conditions, and their fecal samples were collected after two months to assess the gut microbiota by 16S rRNA sequencing.
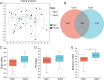
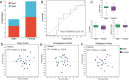
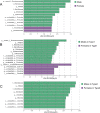
Results: The gut microbiota of captive striped hamsters naturally separated into two types at the amplicon sequence variant (ASV) level. There was a significant difference in the Shannon index among these two types. A receiver operating characteristic (ROC) curve showed that the top 30 ASVs could effectively distinguish each type. Linear discriminant analysis of effect size (LEfSe) showed enrichment of the genera Lactobacillus, Treponema and Pygmaiobacter in one gut microbial type and enrichment of the genera Turicibacter and Ruminiclostridium in the other. The former type had higher carbohydrate metabolism ability, while the latter harbored a more complex co-occurrence network and higher amino acid metabolism ability. The gut microbial types were not associated with sex; however, we did find sex differences in the relative abundances of certain bacterial taxa, including some type-specific sex variations.
Conclusions: Although captive animals live in a unified environment, their gut bacteria can still differentiate into distinct types, but the sex of the hosts may not play an important role in the typing process of small-scale captive animal communities. The relevant driving factors as well as other potential types need to be further investigated to better understand host-microbe interactions.
Keywords: 16S rRNA; Gut microbiota; Rodent; Sex; Typing analysis.
©2023 Fan et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



References
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M’rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

