Molecular dynamics analysis of superoxide dismutase 1 mutations suggests decoupling between mechanisms underlying ALS onset and progression
- PMID: 37954145
- PMCID: PMC10637862
- DOI: 10.1016/j.csbj.2023.09.016
Molecular dynamics analysis of superoxide dismutase 1 mutations suggests decoupling between mechanisms underlying ALS onset and progression
Abstract
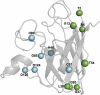
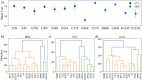
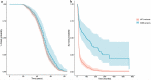
Mutations in the superoxide dismutase 1 (SOD1) gene are the second most common known cause of ALS. SOD1 variants express high phenotypic variability and over 200 have been reported in people with ALS. It was previously proposed that variants can be broadly classified in two groups, 'wild-type like' (WTL) and 'metal binding region' (MBR) variants, based on their structural location and biophysical properties. MBR variants, but not WTL variants, were associated with a reduction of SOD1 enzymatic activity. In this study we used molecular dynamics and large clinical datasets to characterise the differences in the structural and dynamic behaviour of WTL and MBR variants with respect to the wild-type SOD1, and how such differences influence the ALS clinical phenotype. Our study identified marked structural differences, some of which are observed in both variant groups, while others are group specific. Moreover, collecting clinical data of approximately 500 SOD1 ALS patients carrying variants, we showed that the survival time of patients carrying an MBR variant is generally longer (∼6 years median difference, p < 0.001) with respect to patients with a WTL variant. In conclusion, our study highlighted key differences in the dynamic behaviour between WTL and MBR SOD1 variants, and between variants and wild-type SOD1 at an atomic and molecular level, that could be further investigated to explain the associated phenotypic variability. Our results support the hypothesis of a decoupling between mechanisms of onset and progression of SOD1 ALS, and an involvement of loss-of-function of SOD1 with the disease progression.
Keywords: Amyotrophic lateral sclerosis; Diseaseassociated SOD1 mutations; Molecular dynamics (MD) simulations; Performed principal component analysis; Superoxide Dismutase type 1 (SOD1); Survival analysis.
© 2023 Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
None.
Figures






References
-
- Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2(11):806–819. - PubMed
-
- Boillée S., Velde C.V., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52(1):39–59. - PubMed
-
- Perrone B., Conforti F.L. Common mutations of interest in the diagnosis of amyotrophic lateral sclerosis: how common are common mutations in ALS genes? Expert Rev. Mol. Diagn. 2020;20(7):703–714. - PubMed
-
- Stathopulos P.B., et al. Calorimetric analysis of thermodynamic stability and aggregation for apo and holo amyotrophic lateral sclerosis-associated Gly-93 mutants of superoxide dismutase. J Biol Chem. 2006;281(10):6184–6193. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
