Prediction of the interaction between Calloselasma rhodostoma venom-derived peptides and cancer-associated hub proteins: A computational study
- PMID: 37954374
- PMCID: PMC10637925
- DOI: 10.1016/j.heliyon.2023.e21149
Prediction of the interaction between Calloselasma rhodostoma venom-derived peptides and cancer-associated hub proteins: A computational study
Abstract
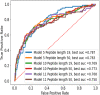
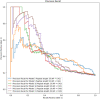
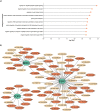
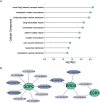
The use of peptide drugs to treat cancer is gaining popularity because of their efficacy, fewer side effects, and several advantages over other properties. Identifying the peptides that interact with cancer proteins is crucial in drug discovery. Several approaches related to predicting peptide-protein interactions have been conducted. However, problems arise due to the high costs of resources and time and the smaller number of studies. This study predicts peptide-protein interactions using Random Forest, XGBoost, and SAE-DNN. Feature extraction is also performed on proteins and peptides using intrinsic disorder, amino acid sequences, physicochemical properties, position-specific assessment matrices, amino acid composition, and dipeptide composition. Results show that all algorithms perform equally well in predicting interactions between peptides derived from venoms and target proteins associated with cancer. However, XGBoost produces the best results with accuracy, precision, and area under the receiver operating characteristic curve of 0.859, 0.663, and 0.697, respectively. The enrichment analysis revealed that peptides from the Calloselasma rhodostoma venom targeted several proteins (ESR1, GOPC, and BRD4) related to cancer.
Keywords: Bioinformatics; Biomedical; Cancer; Deep learning; Peptide; Venom.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Cichonska A., Ravikumar B., Parri E., Timonen S., Pahikkala T., Airola A., Wennerberg K., Rousu J., Aittokallio T. Computational-experimental approach to drug-target interaction mapping: a case study on kinase inhibitors. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005678. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

