Current drugs for HIV-1: from challenges to potential in HIV/AIDS
- PMID: 37954841
- PMCID: PMC10637376
- DOI: 10.3389/fphar.2023.1294966
Current drugs for HIV-1: from challenges to potential in HIV/AIDS
Abstract
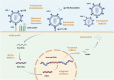
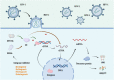
The human immunodeficiency virus (HIV) persists in latently infected CD4+T cells and integrates with the host genome until cell death. Acquired immunodeficiency syndrome (AIDS) is associated with HIV-1. Possibly, treating HIV/AIDS is an essential but challenging clinical goal. This review provides a detailed account of the types and mechanisms of monotherapy and combination therapy against HIV-1 and describes nanoparticle and hydrogel delivery systems. In particular, the recently developed capsid inhibitor (Lenacapavir) and the Ainuovirine/tenofovir disoproxil fumarate/lamivudine combination (ACC008) are described. It is interestingly to note that the lack of the multipass transmembrane proteins serine incorporator 3 (SERINC3) and the multipass transmembrane proteins serine incorporator 5 (SERINC5) may be one of the reasons for the enhanced infectivity of HIV-1. This discovery of SERINC3 and SERINC5 provides new ideas for HIV-1 medication development. Therefore, we believe that in treating AIDS, antiviral medications should be rationally selected for pre-exposure and post-exposure prophylaxis to avoid the emergence of drug resistance. Attention should be paid to the research and development of new drugs to predict HIV mutations as accurately as possible and to develop immune antibodies to provide multiple guarantees for the cure of AIDS.
Keywords: HIV/AIDS treatment; SERINC3; SERINC5; anti-HIV-1 drugs; combination therapy; monotherapy; predict HIV mutations.
Copyright © 2023 Peng, Zong, Wang, Chen, Chen, Peng and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

