A nematode model to evaluate microdeletion phenotype expression
- PMID: 37956108
- PMCID: PMC10849325
- DOI: 10.1093/g3journal/jkad258
A nematode model to evaluate microdeletion phenotype expression
Abstract
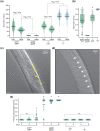
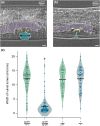
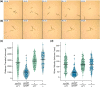
Microdeletion syndromes are genetic diseases caused by multilocus chromosomal deletions too small to be detected by karyotyping. They are typified by complex pleiotropic developmental phenotypes that depend both on the extent of the deletion and variations in genetic background. Microdeletion alleles cause a wide array of consequences involving multiple pathways. How simultaneous haploinsufficiency of numerous adjacent genes leads to complex and variable pleiotropic phenotypes is not well understood. CRISPR/Cas9 genome editing has been shown to induce microdeletion-like alleles at a meaningful rate. Here, we describe a microdeletion allele in Caenorhabditis elegans recovered during a CRISPR/Cas9 genome editing experiment. We mapped the allele to chromosome V, balanced it with a reciprocal translocation crossover suppressor, and precisely defined the breakpoint junction. The allele simultaneously removes 32 protein-coding genes, yet animals homozygous for this mutation are viable as adults. Homozygous animals display a complex phenotype including maternal effect lethality, producing polynucleated embryos that grow into uterine tumors, vulva morphogenesis defects, body wall distensions, uncoordinated movement, and a shortened life span typified by death by bursting. Our work provides an opportunity to explore the complexity and penetrance of microdeletion phenotypes in a simple genetic model system.
Keywords: cytokinesis; genome instability; microdeletion; nematode; phenotype; pleiotropy; sterility.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures



References
-
- Almedom RB, Liewald JF, Hernando G, Schultheis C, Rayes D, Pan J, Schedletzky T, Hutter H, Bouzat C, Gottschalk A. 2009. An ER-resident membrane protein complex regulates nicotinic acetylcholine receptor subunit composition at the synapse. EMBO J. 28(17):2636–2649. doi:10.1038/emboj.2009.204. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
