VarCards2: an integrated genetic and clinical database for ACMG-AMP variant-interpretation guidelines in the human whole genome
- PMID: 37956311
- PMCID: PMC10767961
- DOI: 10.1093/nar/gkad1061
VarCards2: an integrated genetic and clinical database for ACMG-AMP variant-interpretation guidelines in the human whole genome
Abstract
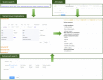
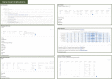
VarCards, an online database, combines comprehensive variant- and gene-level annotation data to streamline genetic counselling for coding variants. Recognising the increasing clinical relevance of non-coding variations, there has been an accelerated development of bioinformatics tools dedicated to interpreting non-coding variations, including single-nucleotide variants and copy number variations. Regrettably, most tools remain as either locally installed databases or command-line tools dispersed across diverse online platforms. Such a landscape poses inconveniences and challenges for genetic counsellors seeking to utilise these resources without advanced bioinformatics expertise. Consequently, we developed VarCards2, which incorporates nearly nine billion artificially generated single-nucleotide variants (including those from mitochondrial DNA) and compiles vital annotation information for genetic counselling based on ACMG-AMP variant-interpretation guidelines. These annotations include (I) functional effects; (II) minor allele frequencies; (III) comprehensive function and pathogenicity predictions covering all potential variants, such as non-synonymous substitutions, non-canonical splicing variants, and non-coding variations and (IV) gene-level information. Furthermore, VarCards2 incorporates 368 820 266 documented short insertions and deletions and 2 773 555 documented copy number variations, complemented by their corresponding annotation and prediction tools. In conclusion, VarCards2, by integrating over 150 variant- and gene-level annotation sources, significantly enhances the efficiency of genetic counselling and can be freely accessed at http://www.genemed.tech/varcards2/.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E.et al. .. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015; 17:405–424. - PMC - PubMed
-
- Elkon R., Agami R.. Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 2017; 35:732–746. - PubMed
MeSH terms
Substances
Grants and funding
- 2021YFC2502100/National Key R&D Program of China
- 32070591/National Natural Science Foundation of China
- 2023JJ30975/Natural Science Foundation of Hunan Province
- 2023SK2093-1]/Scientific Research Program of FuRong Laboratory
- 2023QYJC010/Central South University Research Program of Advanced Interdisciplinary Study
LinkOut - more resources
Full Text Sources

