K-Means Clustering Coarse-Graining (KMC-CG): A Next Generation Methodology for Determining Optimal Coarse-Grained Mappings of Large Biomolecules
- PMID: 37957028
- PMCID: PMC10720621
- DOI: 10.1021/acs.jctc.3c01053
K-Means Clustering Coarse-Graining (KMC-CG): A Next Generation Methodology for Determining Optimal Coarse-Grained Mappings of Large Biomolecules
Abstract
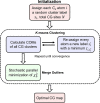
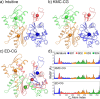
Coarse-grained (CG) molecular dynamics (MD) has become a method of choice for simulating various large scale biomolecular processes; therefore, the systematic definition of the CG mappings for biomolecules remains an important topic. Appropriate CG mappings can significantly enhance the representability of a CG model and improve its ability to capture critical features of large biomolecules. In this work, we present a systematic and more generalized method called K-means clustering coarse-graining (KMC-CG), which builds on the earlier approach of essential dynamics coarse-graining (ED-CG). KMC-CG removes the sequence-dependent constraints of ED-CG, allowing it to explore a more extensive space and thus enabling the discovery of more physically optimal CG mappings. Furthermore, the implementation of the K-means clustering algorithm can variationally optimize the CG mapping with efficiency and stability. This new method is tested in three cases: ATP-bound G-actin, the HIV-1 CA pentamer, and the Arp2/3 complex. In these examples, the CG models generated by KMC-CG are seen to better capture the structural, dynamic, and functional domains. KMC-CG therefore provides a robust and consistent approach to generating CG models of large biomolecules that can then be more accurately parametrized by either bottom-up or top-down CG force fields.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

