Exosome-targeted delivery of METTL14 regulates NFATc1 m6A methylation levels to correct osteoclast-induced bone resorption
- PMID: 37957146
- PMCID: PMC10643436
- DOI: 10.1038/s41419-023-06263-4
Exosome-targeted delivery of METTL14 regulates NFATc1 m6A methylation levels to correct osteoclast-induced bone resorption
Abstract
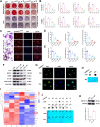
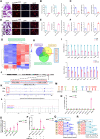
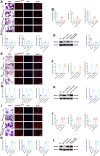
Osteoporosis has a profound influence on public health. First-line bisphosphonates often cause osteonecrosis of the jaw meanwhile inhibiting osteoclasts. Therefore, it is important to develop effective treatments. The results of this study showed that the increased level of NFATc1 m6A methylation caused by zoledronic acid (ZOL), with 4249A as the functional site, is highly correlated with the decreased bone resorption of osteoclasts. Upstream, METTL14 regulates osteoclast bone absorption through the methylation functional site of NFATc1. Downstream, YTHDF1 and YTHDF2 show antagonistic effects on the post-transcriptional regulation of NFATc1 after the m6A methylation level is elevated by METTL14. In this study, meRIP-Seq, luciferase reporter assays, meRIP and other methods were used to elucidate the NFATc1 regulatory mechanism of osteoclasts from the perspective of RNA methylation. In addition, EphA2 overexpression on exosomes is an effective biological method for targeted delivery of METTL14 into osteoclasts. Importantly, this study shows that METTL14 released by exosomes can increase the m6A methylation level of NFATc1 to inhibit osteoclasts, help postmenopausal osteoporosis patients preserve bone mass, and avoid triggering osteonecrosis of the jaw, thus becoming a new bioactive molecule for the treatment of osteoporosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

