Senolytic therapy alleviates physiological human brain aging and COVID-19 neuropathology
- PMID: 37957361
- PMCID: PMC10724067
- DOI: 10.1038/s43587-023-00519-6
Senolytic therapy alleviates physiological human brain aging and COVID-19 neuropathology
Abstract
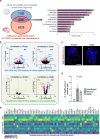
Aging is a major risk factor for neurodegenerative diseases, and coronavirus disease 2019 (COVID-19) is linked to severe neurological manifestations. Senescent cells contribute to brain aging, but the impact of virus-induced senescence on neuropathologies is unknown. Here we show that senescent cells accumulate in aged human brain organoids and that senolytics reduce age-related inflammation and rejuvenate transcriptomic aging clocks. In postmortem brains of patients with severe COVID-19 we observed increased senescent cell accumulation compared with age-matched controls. Exposure of human brain organoids to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induced cellular senescence, and transcriptomic analysis revealed a unique SARS-CoV-2 inflammatory signature. Senolytic treatment of infected brain organoids blocked viral replication and prevented senescence in distinct neuronal populations. In human-ACE2-overexpressing mice, senolytics improved COVID-19 clinical outcomes, promoted dopaminergic neuron survival and alleviated viral and proinflammatory gene expression. Collectively our results demonstrate an important role for cellular senescence in driving brain aging and SARS-CoV-2-induced neuropathology, and a therapeutic benefit of senolytic treatments.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

